Abstract
Objective
Escherichia coli K12f-pACLYC has a high capability for growth and lycopene production when using fructose as carbon source and the transcription of genes involved was compared in glucose-grown and fructose-grown cells.
Results
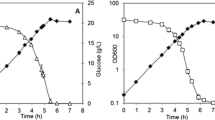
Escherichia coli K12f-pACLYC was grown on 10 g fructose l−1 and reached 4.6 g DCW l−1 with lycopene at 192 mg g DCW−1, values that are 3-fold and 7-fold higher than when growing on glucose. Gene transcription profiles of fructose-grown and glucose-grown cells were compared. 384 differentially expressed genes (DEGs) with fold changes ≥4 were identified, and the transcription of genes involved in fructose uptake and metabolism, pyruvate catabolism, tricarboxylic acid cycle and oxidative phosphorylation varied significantly. These changes enhanced the metabolic flux into the Embden–Meyerhof–Parnas pathway and the tricarboxylic acid cylcle and coupled to oxidative phosphorylation. These enhanced activities provide more precursors, cofactors and energy needed for growth lycopene production.
Conclusion
The high capability of E. coli K12f-pACLYC for growth and lycopene production when growing on fructose is due to transcriptional regulation, and the relevant genes were identified.



Similar content being viewed by others
References
Alper H, Jin YS, Moxley JF, Stephanopoulos G (2005) Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 7:155–164
Cunningham FX Jr, Sun Z, Chamovitz D, Hirschberg J, Gantt E (1994) Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell 6:1107–1121
Farmer WR, Liao JC (2000) Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol 18:533–537
Gallego-Jara J, De Diego T, Del Real A, Ecija-Conesa A et al (2015) Lycopene overproduction and in situ extraction in organic-aqueous culture systems using a metabolically engineered Escherichia coli. AMB Express 5:65
Kim SW, Keasling JD (2001) Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng 72:408–415
Kim SW, Kim JB, Ryu JM, Jung JK et al (2009) High-level production of lycopene in metabolically engineered E. coli. Process Biochem 44:899–905
Lavecchia R, Zuorro A (2008) Improved lycopene extraction from tomato peels using cell-wall degrading enzymes. Eur Food Res and Technol 228:153–158
Lee PC, Schmidt-Dannert C (2002) Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl Microbiol Biotechnol 60:1–11
Sauer U, Canonaco F, Heri S, Perrenoud A et al (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619
Srivastava S, Srivastava AK (2013) Lycopene; chemistry, biosynthesis, metabolism and degradation under various abiotic parameters. Int J Food Sci Tech 52:41–53
Vaishampayan U, Hussain M, Banerjee M, Seren S (2007) Lycopene and soy isoflavones in the treatment of prostate cancer. Nutr Cancer 59:1–7
Yoon SH, Kim JE, Lee SH, Park HM et al (2006) Engineering the lycopene synthetic pathway in E. coli by comparison of the carotenoid genes of Pantoea agglomerans and Pantoea ananatis. Appl Microbiol Biotechnol 74:131–139
Zhao J, Li Q, Sun T, Zhu X et al (2013) Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab Eng 17:42–50
Zhu F, Lu L, Fu S, Zhong XF et al (2015) Targeted engineering and scale up of lycopene overproduction in Escherichia coli. Process Biochem 50:341–346
Acknowledgments
This work was supported by the foundation of the Key Laboratory of Industrial Fermentation Microbiology of the Ministry of Education and the Tianjin Key Lab of Industrial Microbiology (Tianjin University of Science and Technology) (No. 2013IM102).
Supplementary information
Supplementary Table 1—Primers used in quantitative real-time (qRT) PCR analysis.
Supplementary Table 2—Gene ID, name, annotation, and expression pattern and of all the DEGs mentioned involved in the paper.
Supplemetary Fig. 1—COG function classifications of DEGs.
Supplemetary Fig. 2—KEGG function classifications of DEGs.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, W., Song, Y., Liu, M. et al. Gene expression pattern analysis of a recombinant Escherichia coli strain possessing high growth and lycopene production capability when using fructose as carbon source. Biotechnol Lett 38, 1571–1577 (2016). https://doi.org/10.1007/s10529-016-2133-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2133-0