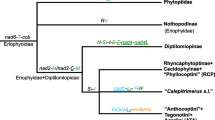
Abstract
The Eriophyoidea, notable for specific morphological characters (four-legged mites) and gall-formation in host plants (gall mites), is one of the most species-rich superfamilies of Acari. Monophyly of the superfamily Eriophyoidea is accepted by all acarologists; however, monophyly of most genera has not been evaluated in a molecular phylogenetic network. Furthermore, most eriophyoid mites, especially species in the genus Diptilomiopus, are morphologically similar, challenging their identification. Here we test the phylogeny and cryptic diversity of Diptilomiopus species using fragments of two mitochondrial (COI and 12S) and two nuclear (18S and 28S) genes. Our results revealed the monophyly of Diptilomiopus. Sequence distance, barcode gap, and species delimitation analyses of the COI gene allowed us to resolve cryptic diversity of Diptilomiopus species. Additionally, we supposed that characteristics of genu fused with femur on both legs and seta ft′ absent on leg II evolved only once within Diptilomiopus, which are potential morphological synapomorphies. In contrast, characteristics of both setae ft′ and ft″ divided into a short branch and a long branch were supposed evolving multiple times independently. Our findings contribute to the understanding of phylogeny and morphological evolution of Diptilomiopus species and provide a DNA-based approach for species delimitation of Diptilomiopus mites.







Similar content being viewed by others

References
Alfaro ME (2003) Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol 20:255–266. https://doi.org/10.1093/molbev/msg028
Amrine JW Jr, Manson DCM (1996) Preparation, mounting and descriptive study of eriophyoid mites. In: Lindquist EE, Sabelis MW, Bruin J (eds) Eriophyoid mites: their biology, natural enemies and control. World Crop Pests, vol 6. Elsevier, Amsterdam, pp 383–396
Amrine JW Jr, Stasny TA, Flechtmann CHW (2003) Revised keys to world genera of Eriophyoidea (Acari: Prostigmata). Indira Publishing House, Michigan
Amrine JW Jr, Stasny TA (1994) Catalog of the Eriophyoidea (Acarina: Prostigmata) of the world. Indira Publishing House, Michigan
Campbell CL, Tanaka N, White KH, Thorsness PE (1994) Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell 5:899–905. https://doi.org/10.1091/mbc.5.8.899
Capinera JL (2008) Encyclopedia of entomology. Springer, Dordrecht
Chandrapatya A, Konvipasruang P, Amrine JW Jr (2016) New eriophyoid mites from Thailand. Syst Appl Acarol 21:55–78. https://doi.org/10.11158/saa.21.1.5
Chetverikov PE, Craemer C, Neser S (2018) New pseudotagmic genus of acaricaline mites (Eriophyidae, Acaricalini) from a South African palm Hyphaene coriacea and remarks on lateral opisthosomal spines and morphology of deutogynes in Eriophyoidea. Syst Appl Acarol 23:1073–1101. https://doi.org/10.11158/saa.23.6.6
Chetverikov PE, Cvrkovic T, Makunin A, Sukhareva S, Vidovic B, Petanovic R (2015) Basal divergence of Eriophyoidea (Acariformes, Eupodina) inferred from combined partial COI and 28S gene sequences and CLSM genital anatomy. Exp Appl Acarol 67:219–245. https://doi.org/10.1007/s10493-015-9945-9
Craemer C, Amrine JW Jr, Childers CC, Rogers ME, Achor DS (2017) A new eriophyoid mite species, Diptilomiopus floridanus (Acari: Eriophyoidea: Diptilomiopidae), from citrus in Florida, USA. Syst Appl Acarol 22:386–402. https://doi.org/10.11158/saa.22.3.5
Cvrković T, Chetverikov P, Vidović B, Petanović R (2016) Cryptic speciation within Phytoptus avellanae s.l. (Eriophyoidea: Phytoptidae) revealed by molecular data and observations on molting Tegonotus-like nymphs. Exp Appl Acarol 68:83–96. https://doi.org/10.1007/s10493-015-9981-5
Dabert J, Ehrnsberger R, Dabert M (2008) Glaucalges tytonis sp. n. (Analgoidea, Xolalgidae) from the barn owl Tyto alba (Strigiformes, Tytonidae): compiling morphology with DNA barcode data for taxon descriptions in mites (Acari). Zootaxa 1719:41–52
Dabert M, Witalinski W, Kazmierski A, Olszanowski Z, Dabert J (2010) Molecular phylogeny of acariform mites (Acari, Arachnida): strong conflict between phylogenetic signal and long-branch attraction artifacts. Mol Phylogenet Evol 56:222–241. https://doi.org/10.1016/j.ympev.2009.12.020
Duarte ME, de Mendonca RS, Skoracka A, Silva ES, Navia D (2019) Integrative taxonomy of Abacarus mites (Eriophyidae) associated with hybrid sugarcane plants, including description of a new species. Exp Appl Acarol 78: 373–401. https://doi.org/10.1007/s10493-019-00388-y
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. https://doi.org/10.1186/1471-2148-7-214
Ezard THG, Fujisawa TE, Barraclough TG (2014) Splits: species' limits by threshold statistics. R package version 1.0–19/r51. http://RForge.Rproject.org/projects/splits/
Flechtmann CHW (2004) Eriophyid mites (AcariL Eriophyidae) from brazilian sedges (Cyperaceae). Int J Acarol 30:157–164. https://doi.org/10.1080/01647950408684385
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol 62:707–724. https://doi.org/10.1093/sysbio/syt033
Guo J-F, Li H-S, Wang B, Xue X-F, Hong X-Y (2015) DNA barcoding reveals the protogyne and deutogyne of Tegolophus celtis sp. nov. (Acari: Eriophyidae). Exp Appl Acarol 67:393–410. https://doi.org/10.1007/s10493-015-9953-9
Hillis DM, Dixon MT (1991) Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 66:411–453. https://doi.org/10.1086/417338
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192. https://doi.org/10.1093/sysbio/42.2.182
Huang K-W, Wang C-F (2004) Eriophyoid mites of Taiwan: descriptin of three species of Calacarini from Hueysuen (Acari: Eriophyoidea: Phyllocoptinae). Zootaxa 527:1–8
Kambhampati S, Smith PT (1995) PCR primers for the amplification of four insect mitochondrial gene fragments. Insect Mol Biol 4:233–236. https://doi.org/10.1111/j.1365-2583.1995.tb00028.x
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Klimov PB, OConnor BM, Chetverikov PE, Bolton SJ, Pepato AR, Mortazavi AL, Tolstikov AV, Bauchan GR, Ochoa R (2018) Comprehensive phylogeny of acariform mites (Acariformes) provides insights on the origin of the four-legged mites (Eriophyoidea), a long branch. Mol Phylogenet Evol 119:105–117. https://doi.org/10.1016/j.ympev.2017.10.017
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772–773. https://doi.org/10.1093/molbev/msw260
Lewandowski M, Skoracka A, Szydlo W, Kozak M, Druciarek T, Griffiths DA (2014) Genetic and morphological diversity of Trisetacus species (Eriophyoidea: Phytoptidae) associated with coniferous trees in Poland: phylogeny, barcoding, host and habitat specialization. Exp Appl Acarol 63:497–520. https://doi.org/10.1007/s10493-014-9805-z
Li H-S, Xue X-F, Hong X-Y (2014a) Homoplastic evolution and host association of Eriophyoidea (Acari, Prostigmata) conflict with the morphological-based taxonomic system. Mol Phylogenet Evol 78:185–198. https://doi.org/10.1016/j.ympev.2014.05.014
Li H-S, Xue X-F, Hong X-Y (2014b) Cryptic diversity in host-associated populations of Tetra pinnatifidae (Acari: Eriophyoidea): What do morphometric, mitochondrial and nuclear data reveal and conceal? Bull Entomol Res 104:221–232. https://doi.org/10.1017/S0007485313000746
Lindquist EE (1996) External anatomy and notation of structures. In: Lindquist EE, Sabelis MW, Bruin J (eds) Eriophyoid mites: their biology, natural enemies and control. World Crop Pests, vol 6. Elsevier, Amsterdam, pp 3–31
Liu S, Li J, Guo K, Qiao H, Xu R, Chen J, Xu C, Chen J (2016) Seasonal phoresy as an overwintering strategy of a phytophagous mite. Sci Rep 6:25483. https://doi.org/10.1038/srep25483
Michalska K, Skoracka A, Navia D, Amrine JW Jr (2010) Behavioural studies on eriophyoid mites: an overview. Exp Appl Acarol 51:31–59. https://doi.org/10.1007/s10493-009-9319-2
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov 2010, New Orleans, pp 1–8
Nalepa A (1916) Neue Gallmilben (32. Fortsetzung). Anzeiger der kaiserlichen Akademie Wissenschaften. Mathematische-naturwissenschaftliche Klasse Wien 53:283–284
Navia D, de Mendonca RS, Skoracka A, Szydlo W, Knihinicki D, Hein GL, da Silva Pereira PR, Truol G, Lau D (2013) Wheat curl mite, Aceria tosichella, and transmitted viruses: an expanding pest complex affecting cereal crops. Exp Appl Acarol 59:95–143. https://doi.org/10.1007/s10493-012-9633-y
Petanovic R, Kielkiewicz M (2010) Plant-eriophyoid mite interactions: specific and unspecific morphological alterations. Part II. Exp Appl Acarol 51:81–91. https://doi.org/10.1007/s10493-009-9328-1
Pons J, Barraclough T, Gomez-Zurita J, Cardoso A, Duran D, Hazell S, Kamoun S, Sumlin W, Vogler A (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55:595–609. https://doi.org/10.1080/10635150600852011
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol 21:1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Sabelis MW, Bruin J (1996) Evolutionary ecology: Life history patterns, food plant choice and dispersal. In: Lindquist EE, Sabelis MW, Bruin J (eds) Eriophyoid mites: their biology, natural enemies and control. World Crop Pests, vol 6. Elsevier, Amsterdam, pp 329–366
Schmidt AR, Jancke S, Lindquist EE, Ragazzi E, Roghi G, Nascimbene PC, Schmidt K, Wappler T, Grimaldi DA (2012) Arthropods in amber from the Triassic Period. Proc Natl Acad Sci USA 109:14796–14801. https://doi.org/10.1073/pnas.1208464109
Sidorchuk EA, Schmidt AR, Ragazzi E, Roghi G, Lindquist EE (2015) Plant-feeding mite diversity in Triassic amber (Acari: Tetrapodili). J Syst Palaeontol 13:129–151. https://doi.org/10.1080/14772019.2013.867373
Silvestro D, Michalak I (2011) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Skoracka A, Dabert M (2010) The cereal rust mite Abacarus hystrix (Acari: Eriophyoidea) is a complex of species: evidence from mitochondrial and nuclear DNA sequences. Bull Entomol Res 100:263–272. https://doi.org/10.1017/S0007485309990216
Skoracka A, Kuczyński L, Szydło W, Rector B (2013) The wheat curl mite Aceria tosichella (Acari: Eriophyoidea) is a complex of cryptic lineages with divergent host ranges: evidence from molecular and plant bioassay data. Biol J Linnean Soc 109:165–180. https://doi.org/10.1111/bij.12024
Skoracka A, Lopes LF, Alves MJ, Miller A, Lewandowski M, Szydlo W, Majer A, Rozanska E, Kuczynski L (2018) Genetics of lineage diversification and the evolution of host usage in the economically important wheat curl mite, Aceria tosichella Keifer, 1969. BMC Evol Biol 18:122. https://doi.org/10.1186/s12862-018-1234-x
Skoracka A, Smith L, Oldfield G, Cristofaro M, Amrine JW Jr (2010) Host-plant specificity and specialization in eriophyoid mites and their importance for the use of eriophyoid mites as biocontrol agents of weeds. Exp Appl Acarol 51:93–113. https://doi.org/10.1007/s10493-009-9323-6
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Toews DP, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21:3907–3930. https://doi.org/10.1111/j.1365-294X.2012.05664.x
Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC (1997) The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol 46:1–68. https://doi.org/10.1093/sysbio/46.1.1
Xue X-F, Song Z-W, Amrine JW Jr, Hong X-Y (2007) Eriophyoid mites on coniferous plants in China with descriptions of a new genus and five new species (Acari: Eriophyoidea). Int J Acarol 33:333–345. https://doi.org/10.1080/01647950708683695
Xue X-F, Dong Y, Deng W, Hong X-Y, Shao R (2017) The phylogenetic position of eriophyoid mites (superfamily Eriophyoidea) in Acariformes inferred from the sequences of mitochondrial genomes and nuclear small subunit (18S) rRNA gene. Mol Phylogenet Evol 109:271–282. https://doi.org/10.1016/j.ympev.2017.01.009
Xue X-F, Guo J-F, Dong Y, Hong X-Y, Shao R (2016) Mitochondrial genome evolution and tRNA truncation in Acariformes mites: new evidence from eriophyoid mites. Sci Rep 6:18920. https://doi.org/10.1038/srep18920
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. https://doi.org/10.1093/bioinformatics/btt499
Zhang Z-Q (2011) Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Magnolia Press, Auckland
Zhang Z-Q (2018) Repositories for mite and tick specimens: acronyms and their nomenclature. Syst Appl Acarol 23:2432–2466. https://doi.org/10.11158/saa.23.12.12
Zhao S, Amrine JW Jr (1997) Investigation of snowborne mites (Acari) and relevancy to dispersal. Int J Acarol 23:209–213. https://doi.org/10.1080/01647959708683565
Zhao Y, Li W, Wang G-Q (2018) New asian phyllocoptines (Eriophyidae, Phyllocoptinae): descriptions of Namengia latifloris gen. nov. & sp. nov. (Acaricalini) and Petanovicia cathartica sp. nov. (Phyllocoptini) from northwest Laos. Syst Appl Acarol 23:2022–2032. https://doi.org/10.11158/saa.23.10.11
Acknowledgements
We would like to thank Mr. Jimmy Chew (Borneo Jungle Girl Camp, Keningau, Sabah, Malaysia) for kindly collecting mite specimens in Sabah, Malaysia. This research was funded by the National Natural Science Foundation of China (31970437).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
10493_2019_443_MOESM1_ESM.pdf
Supplementary material 1 Phylogenetic trees inferred from nucleotide sequences of 18S gene using maximum likelihood method. Node numbers indicate maximum likelihood bootstrap proportion (BSP) (PDF 166 kb)
10493_2019_443_MOESM2_ESM.pdf
Supplementary material 2 Phylogenetic trees inferred from nucleotide sequences of 18S gene using Bayesian method. Node numbers indicate Bayesian posterior probabilities (BPP) (PDF 159 kb)
10493_2019_443_MOESM3_ESM.pdf
Supplementary material 3 Phylogenetic trees inferred from nucleotide sequences of two mitochondrial (COI and 12S) and two nuclear (18S and 28S) gene fragments using maximum likelihood method. Node numbers indicate maximum likelihood bootstrap proportion (BSP) (135 kb)
10493_2019_443_MOESM4_ESM.pdf
Supplementary material 4 Phylogenetic trees inferred from nucleotide sequences of two mitochondrial (COI and 12S) and two nuclear (18S and 28S) gene fragments using Bayesian method. Node numbers indicate Bayesian posterior probabilities (BPP) (134 kb)
10493_2019_443_MOESM5_ESM.pdf
Supplementary material 5 Phylogenetic trees of Diptilomiopus species inferred from nucleotide sequences of (A) COI (no partitioned), (B) COI (partitioned by three codons), (C) 18S and (D) 28S using Bayesian method. Node numbers (black) indicate Bayesian posterior probabilities (BPP). Branch numbers (red) indicate support values inferred by bPTP. Blue bars indicate delimitated species by analyses of GYMC and bPTP (440 kb)
Rights and permissions
About this article
Cite this article
Liu, Q., Yuan, YM., Lai, Y. et al. Unravelling the phylogeny, cryptic diversity and morphological evolution of Diptilomiopus mites (Acari: Eriophyoidea). Exp Appl Acarol 79, 323–344 (2019). https://doi.org/10.1007/s10493-019-00443-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-019-00443-8