Abstract
We describe the species Metschnikowia bowlesiae sp. nov. based on the recovery of six isolates from Hawaii and Belize. The species belongs to the Metschnikowia arizonensis subclade of the large-spored Metschnikowia clade. The isolates are haploid and heterothallic. Both Hawaiian strains had the mating type h + and the Belizean strains were h −. Paraphyletic species structures observed in some ribosomal DNA sequence analyses suggest that M. bowlesiae sp. nov. might represent an intermediate stage in a succession of peripatric speciation events from Metschnikowia dekortorum to Metschnikowia similis and might even hybridize with these species. The type of M. bowlesiae sp. nov. is strain UWOPS 04-243x5 (CBS 12940T, NRRL Y-63671) and the allotype is strain UWOPS 12-619.1 (CBS 12939A, NRRL Y-63670).



Similar content being viewed by others
References
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Fidalgo-Jiménez A, Daniel HM, Evrard P, Decock C, Lachance MA (2008) Metschnikowia cubensis sp. nov., a new yeast species isolated from flowers in Cuba. Int J Syst Evol Microbiol 58:2955–2961
Guzmán B, Lachance MA, Herrera CM (2013) Phylogenetic analysis of the angiosperm–floricolous insect–yeast association: have yeast and angiosperm lineages co-diversified? Mol Phylogenet Evol 68:161–175
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for the isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, vol 1. Elsevier, Amsterdam, pp 87–110
Lachance MA (2011) Metschnikowia Kamienski (1899). In: Kurtzman CP, Fell JW, Boekhout T (eds) The Yeasts, a taxonomic study, vol 1. Elsevier, Amsterdam, pp 575–620
Lachance MA, Bowles JM (2002) Metschnikowia arizonensis and Metschnikowia dekortorum, two new large-spored yeast species associated with floricolous beetles. FEMS Yeast Res 2:81–86
Lachance MA, Bowles JM (2004) Metschnikowia similis sp. nov. and Metschnikowia colocasiae sp.nov., two ascomycetous yeasts isolated from Conotelus spp. (Coleoptera: Nitidulidae) in Costa Rica. Stud Mycol 50:69–76
Lachance MA, Starmer WT, Phaff HG (1990) Metschnikowia hawaiiensis sp. nov., a heterothallic haploid yeast from hawaiian morning glory and associated drosophilids. Int J Syst Bacteriol 40:415–420
Lachance MA, Starmer WT, Bowles JM, Phaff HJ, Rosa CA (2000) Ribosomal DNA, species structure, and biogeography of the cactophilic yeast Clavispora opuntiae. Can J Microbiol 46:195–210
Lachance MA, Kaden JE, Phaff HJ, Starmer WT (2001) Phylogenetic structure of the Sporopachydermia cereana species complex. Int J Syst Evol Microbiol 51:237–247
Lachance MA, Bowles JM, Starmer WT (2003a) Geography and niche occupancy as determinants of yeast biodiversity: the yeast–insect–morning glory ecosystem of Kīpuka Puaulu, Hawai’i. FEMS Yeast Res 4:104–111
Lachance MA, Daniel HM, Meyer W, Prasad GS, Gautam SP, Boundy-Mills K (2003b) The D1/D2 domain of the large-subunit rDNA of the yeast species Clavispora lusitaniae is unusually polymorphic. FEMS Yeast Res 4:253–258
Lachance MA, Ewing CP, Bowles JM, Starmer WT (2005) Metschnikowia hamakuensis sp. nov., Metschnikowia kamakouana sp. nov., and Metschnikowia mauinuiana sp. nov., three endemic yeasts from Hawaiian nitidulid beetles. Int J Syst Evol Microbiol 55:1369–1377
Marinoni G, Lachance MA (2004) Speciation in the large-spored Metschnikowia clade and establishment of a new species, Metschnikowia borealis comb. nov. FEMS Yeast Res 4:587–596
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Rosa CA, Lachance MA, Teixeira LCRS, Pimenta RS, Morais PB (2007) Metschnikowia cerradonensis sp. nov., a yeast species isolated from ephemeral flowers and their nitidulid beetles in Brazil. Int J Syst Evol Microbiol 57:161–165
Russell J, Zomerdijk JCBM (2005) RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30:87–96
Suchard MA, Weiss RE, Dorman KS, Sinsheimer JS (2003) Inferring spatial phylogenetic variation along nucleotide sequences: a multiple change-point model. J Am Stat Assoc 98:427–437
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgments
This work was funded by the Natural Science and Engineering Council of Canada. We acknowledge the Granting of research or collecting permits by the Department of Land and Natural Resources of Hawaii, USA, and the Ministry of Natural Resources and the Environment of Belize. Field assistance from Jane Bowles and Curtis Ewing is gratefully acknowledged. The work in Belize was facilitated by Brock Fenton and Mark Howells. We thank Sheila Macfie for her critical reading of the revised manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
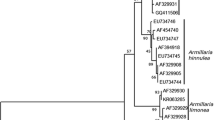
Phylogram of representatives of each ITS-D1/D2 phylotype (Fig. 2) of M. bowlesiae sp. nov., M. dekortorum, and M. similis with M. colocasiae as outgroup. The neighbour-joining analysis was performed on 1946 aligned positions in the ribosomal intergenic spacer (IGS). Gapped positions were deleted on a pair-wise basis. The scale bar shows the number of substitutions, which was the distance measure used in the analysis. Bootstraps from 1,000 replicates are shown. Supplementary material 1 (PDF 184 kb)
Online Resource 2
Phylogram showing the placement of M. bowlesiae sp. nov. in the M. arizonensis subclade. A maximum likelihood analysis was performed on 5439 non-gapped, aligned positions in the ribosomal gene cluster. The distances were fit to the general time reversible model. Bootstraps for 100 iterations are shown. A discrete Γ distribution was used to accommodate rate differences among sites (5 categories, Γ parameter = 0.05). The scale shows the patristic distance. Supplementary material 2 (PDF 102 kb)
Rights and permissions
About this article
Cite this article
Lachance, MA., Fedor, A.N. Catching speciation in the act: Metschnikowia bowlesiae sp. nov., a yeast species found in nitidulid beetles of Hawaii and Belize. Antonie van Leeuwenhoek 105, 541–550 (2014). https://doi.org/10.1007/s10482-013-0106-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-0106-z