Abstract
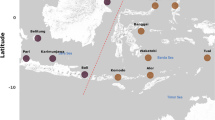
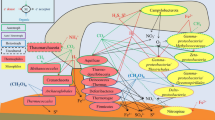
This study was conducted to characterize the diversity of microbial communities in marine sediments of the South China Sea by means of 16S rRNA gene clone libraries. The results revealed that the sediment samples collected in summer harboured a more diverse microbial community than that collected in winter, Deltaproteobacteria dominated 16S rRNA gene clone libraries from both seasons, followed by Gammaproteobacteria, Acidobacteria, Nitrospirae, Planctomycetes, Firmicutes. Archaea phylotypes were also found. The majority of clone sequences shared greatest similarity to uncultured organisms, mainly from hydrothermal sediments and cold seep sediments. In addition, the sedimentary microbial communities in the coastal sea appears to be much more diverse than that of the open sea. A spatial pattern in the sediment samples was observed that the sediment samples collected from the coastal sea and the open sea clustered separately, a novel microbial community dominated the open sea. The data indicate that changes in environmental conditions are accompanied by significant variations in diversity of microbial communities at the South China Sea.





Similar content being viewed by others
References
Acinas SG, Antón J, Rodríguez-Valera F (1999) Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol 65:514–522
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Baker BJ, Banfield JF (2003) Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152
Bowman JP, McCuaig RD (2003) Biodiversity, community structural shifts, and biogeography of prokaryotes with Antarctic continental shelf sediment. Appl Environ Microbiol 69:2463–2483
Bruce T, Martinez IB, Maia Neto O, Vicente AC, Kruger RH, Thompson FL (2010) Bacterial community diversity in the Brazilian Atlantic forest soils. Microb Ecol 60:840–849
Copley J (2002) All at sea. Nature 415:572–574
Dang HY, Zhang XX, Sun J, Li TG, Zhang ZN, Yang GP (2008) Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology 154:2084–2095
Deming JW (1998) Deep ocean environmental biotechnology. Curr Opin Biotechnol 9:283–287
Dias AC, Andreote FD, Rigonato J, Fiore MF, Melo IS, Araújo WL (2010) The bacterial diversity in a Brazilian non-disturbed mangrove sediment. Antonie Van Leeuwenhoek. 98:541–551
Fang J, Shizuka A, Kato C, Schouten S (2006) Microbial diversity of cold-seep sediments in Sagami Bay, Japan, as determined by 16S rRNA gene and lipid analyses. FEMS Microbiol Ecol 57:429–441
Fenchel T (2001) Ecology—marine bugs and carbon flow. Science 292:2444–2445
Feng BW, Li XR, Wang JH, Hu ZY, Meng H, Xiang LY (2009) Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea. FEMS Microbiol Ecol 70:80–92
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102:14683–14688
Fuhrman JA, Davis AA (1997) Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNAgene sequences. Mar Ecol Prog Ser 150:275–285
Fuhrman JA, McCallum K, Davis AA (1993) Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol 59:1294–1302
Fukunaga Y, Kurahashi M, Yanagi K, Yokota A, Harayama S (2008) Acanthopleuribacter pedis gen. nov., sp. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagaceae fam. nov., Holophagales ord. nov. and Holophagae classis nov. in the phylum ‘Acidobacteria’. Int J Syst Evol Microbiol 58:2597–2601
Ghosal D, Chakraborty J, Khara P, Dutta TK (2010) Degradation of phenanthrene via meta-cleavage of 2-hydroxy-1-naphthoic acid by Ochrobactrum sp. strain PWTJD. FEMS Microbiol Lett 313:103–110
Giovannoni SJ, Rappé MS (2000) Evolution, diversity and molecular ecology of marine prokaryotes. In: Kirchman DL (ed) Microbial ecology of the oceans. Wiley-Liss, New York, pp 47–84
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60–63
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM (2006) Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:e95
Heijs SK, Laverman AM, Forney LJ, Hardoim PR, van Elsas JD (2008) Comparison of deep-sea sediment microbial communities in the Eastern Mediterranean. FEMS Microbiol Ecol 64:362–377
Huang Y, Lai X, He X, Cao L, Zeng Z, Zhang J, Zhou S (2009) Characterization of a deep-sea sediment metagenomic clone that produces water-soluble melanin in Escherichia coli. Mar Biotechnol (NY) 11:124–131
Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K (2003) Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl Environ Microbiol 69:7224–7235
Jiang L, Zheng Y, Chen J, Xiao X, Wang F (2011) Stratification of Archaeal communities in shallow sediments of the Pearl River Estuary, Southern China. Antonie van Leeuwenhoek. doi:10.1007/s10482-011-9548-3
Karl DM (2002) Hidden in a sea of microbes. Nature 415:590–591
Kelly KM, Chistoserdov AY (2001) Phylogenetic analysis of the succession of bacterial communities in the Great South Bay (Long Island). FEMS Microbiol Ecol 35:85–95
Kishimoto N, Kosako Y, Tano T (1991) Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr Microbiol 22:1–7
Koch IH, Gich F, Dunfield PF, Overmann J (2008) Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int J Syst Evol Microbiol 58:1114–1122
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lai X, Cao L, Tan H, Fang S, Huang Y, Zhou S (2007) Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J 1:756–762
Lebuhn M, Achouak W, Schloter M, Berge O, Meier H, Barakat M, Hartmann A, Heulin T (2000) Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int J Syst Evol Microbiol 50:2207–2223
Li L, Kato C, Horikoshi K (1999) Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol 1:391–400
Liesack W, Bak F, Kreft J-U, Stackebrandt E (1994) Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol 162:85–90
López-García P, Duperron S, Philippot P, Foriel J, Susini J, Moreira D (2003) Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ Microbiol 5:961–976
Madigan MT, Martinko JM, Parker J (1997) Brock biology of microorganisms. Prentice Hall, New Jersey
Martinez RJ, Mills HJ, Story S, Sobecky PA (2006) Prokaryotic diversity and metabolically active microbial populations in sediments from an active mud volcano in the Gulf of Mexico. Environ Microbiol 8:1783–1796
Mills HJ, Hodges C, Wilson K, Macdonald IR, Sobecky PA (2003) Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol Ecol 46:39–52
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810
Norris TB, Wraith JM, Castenholz RW (2002) Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl Environ Microbiol 68:6300–6309
North NN, Dollhopf SL, Petrie L, Istok JD, Balkwill DL, Kostka JE (2004) Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl Environ Microbiol 70:4911–4920
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740
Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA (2010) Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol Ecol 72:429–444
Ravenschlag K, Sahm K, Amann R (2001) Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl Environ Microb 67:387–395
Sapp M, Parker ER, Teal LR, Schratzberger M (2010) Advancing the understanding of biogeography–diversity relationships of benthic microorganisms in the North Sea. FEMS Microbiol Ecol 74:410–429
Sayeh R, Birrien JL, Alain K, Barbier G, Hamdi M, Prieur D (2010) Microbial diversity in Tunisian geothermal springs as detected by molecular and culture-based approaches. Extremophiles 14:501–514
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB (2009) Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Sekiguchi H, Koshikawa H, Hiroki M, Murakami S, Xu K, Watanabe M (2002) Bacterial distribution and phylogenetic diversity in the Changjiang estuary before the construction of the Three Gorges Dam. Microb Ecol 43:82–91
Streit WR, Daniel R, Jaeger KE (2004) Prospecting for biocatalysts and drugs in the genomes of non-cultured microorganisms. Curr Opin Biotechnol 15:285–290
Tan GL, Shu WS, Hallberg KB, Li F, Lan CY, Huang LN (2007) Cultivation-dependent and cultivation-independent characterization of the microbial community in acid mine drainage associated with acidic Pb/Zn mine tailings at Lechang, Guangdong, China. FEMS Microbiol Ecol 59:118–126
Teske A, Brinkhoff T, Muyzer G, Moser DP, Rethmeier J, Jannasch HW (2000) Diversity of thiosulfateoxidizing bacteria from marine sediments and hydrothermal vents. Appl Environ Microbiol 66:3125–3133
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian F, Yu Y, Chen B, Li HR, Yao YF, Guo XK (2009) Bacterial, archaeal and eukaryotic diversity in Arctic sediment as revealed by 16S rRNA and 18S rRNA gene clone libraries analysis. Polar Biol 32:93–103
Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, Giovannoni SJ (2009) Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3:1148–1163
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang P, Li T, Hu A, Wei Y, Guo W, Jiao N, Zhang C (2010) Community structure of archaea from deep-sea sediments of the South China Sea. Microb Ecol 60:796–806
Ward DM, Ferris MJ, Nold SC, Bateson MM (1998) A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev 62:1353–1370
Xu M, Wang P, Wang F, Xiao X (2004) Microbial diversity in deep-sea sediments collected from different stations of Pacific Warm Pool. Mar Biotechnol 6:S161–S167
Xu M, Wang P, Wang F, Xiao X (2005) Microbial diversity at a deep-sea station of the Pacific Nodule Province. Biodivers Conserv 14:3363–3380
Yao B (2005) The forming condition and distribution characteristics of the gas hydrate in the South China Sea. Mar Geol Quat Geol 25:81–90
Zhou MY, Chen XL, Zhao HL, Dang HY, Luan XW, Zhang YZ (2009) Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China. Microb Ecol 58:582–590
Acknowledgments
This research was supported by grants from the National High-Tech Research & Development program of China (2007AA091904 & 2008AA09Z402).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Du, J., Xiao, K., Huang, Y. et al. Seasonal and spatial diversity of microbial communities in marine sediments of the South China Sea. Antonie van Leeuwenhoek 100, 317–331 (2011). https://doi.org/10.1007/s10482-011-9587-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-011-9587-9