Abstract
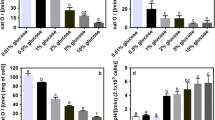
Conventional complex media are routinely used to grow auxotrophic strains under the assumption that they can compensate the latter’s nutritional deficiencies. We here demonstrate that this is not always true. This study compares the growth parameters of Saccharomyces cerevisiae (S288C) and its derived auxotrophic strains FY1679-14C and BY4741 in synthetic minimal medium (SD), standard YPD medium from two of the most commonly used suppliers, or modified YPD medium. Maximum specific growth rates of auxotrophic strains were slightly lower than the prototrophic case in all growth conditions tested. Also, the biomass production of auxotrophic strains in synthetic medium was slightly less than the prototrophic case. However in both of the two standard YPD media used, the biomass production of both auxotrophic strains was markedly lower than that of the prototrophic one. The extent of the differences depended on the medium used. Indeed in one of the two YPD media, the lower biomass production of auxotrophic strains was evident even at the diauxic shift. Uracil seems to be the main limiting growth factor for both auxotrophic strains growing in the two standard YPD medium tested. No YPD media or specific supplement was able to compensate for the effect of the auxotrophic mutations in the multiple auxotrophic marker strain BY4741. The fact that auxotrophic strains grew poorly on YPD when compared to their prototrophic counterpart indicates that standard YPD medium is not sufficient to overcome the effect of auxotrophic mutations.



Similar content being viewed by others
References
Adams A, Gottschling DE, Kaiser CA, Steam T (1998) Methods in Yeast genetics: a cold spring harbor laboratory course manual. Cold Spring Harbor Laboratory Press, NY
Baracat-Pereira M, Coelho J, Minussi R, Chaves-Alves V, Brandão R, Silva D (1999) Cyclic AMP and low molecular weight effector(s) present in yeast extract are involved in pectin lyase production by Penicillium griseoroseum cultured on sucrose. Appl Biochem Biotechnol 76(2):129–141
Botstein D, Davis R (1982) Principles and practice of recombinant DNA research with yeast. In: Strathern JN, Jones EW, Broach JR (eds) The molecular biology of the yeast Saccharomyces cerevisiae. Metabolism and gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp 607–636
Brachmann CB, Davies AJ, Cost G, Caputo E, Li J, Hieter PD, Boeke J (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2):115–132
Çakar ZP, Sauer U, Bailey JE (1999) Metabolic engineering of yeast: the perils of auxotrophic hosts. Biotechnol Lett 21(7):611–616
Çakar ZP, Sauer U, Bailey JE, Müller M, Stolz M, Wallimann T, Schlattner U (2000) Vacuolar morphology and cell cycle distribution are modified by leucine limitation in auxotrophic Saccharomyces cerevisiae. Biol Cell 92(8–9):629–637. doi:10.1016/s0248-4900(01)01111-x
Chopra R, Sharma VM, Ganesan K (1999) Elevated growth of Saccharomyces cerevisiae ATH1 null mutants on glucose is an artifact of nonmatching auxotrophies of mutant and reference strains. Appl Environ Microbiol 65(5):2267–2268
Cohen R, Engelberg D (2007) Commonly used Saccharomyces cerevisiae strains (e.g. BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiol Lett 273(2):239–243
Corbacho I, Olivero I, Hernández LM (2004) Identification of low-dye-binding (ldb) mutants of Saccharomyces cerevisiae. FEMS Yeast Res 4(4–5):437–444
Corbacho I, Olivero I, Hernández LM (2005) A genome-wide screen for Saccharomyces cerevisiae nonessential genes involved in mannosyl phosphate transfer to mannoprotein-linked oligosaccharides. Fungal Genet Biol 42(9):773–790
Crous JM, Pretorius IS, van Zyl WH (1996) Cloning and expression of the α–l -arabinofuranosidase gene (ABF 2) of Aspergillus niger in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 46(3):256–260
Entian K-D, Kötter P (1998) Yeast mutant and plasmid collection. In: Brown JPA, Tuite MF (eds) Methods in Microbiology, vol 36. Elsevier B.V, Amsterdam, pp 431–449
Goldstein AL H, McCusker J (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15(14):1541–1553
Görgens JF, Planas JH, van Zyl WH, Knoetze J, Hahn-Hägerdal B (2004) Comparison of three expression systems for heterologous xylanase production by S cerevisiae in defined medium. Yeast 21(14):1205–1217
Hahn-Hagerdal B, Karhumaa K, Larsson C, Gorwa-Grauslund M, Gorgens J, van Zyl W (2005) Role of cultivation media in the development of yeast strains for large scale industrial use. Microbial Cell Factories 4(1):31
Harsch MJ, Soon AL, Goddard MR, Gardner RC (2009) Optimized fermentation of grape juice by laboratory strains of Saccharomyces cerevisiae. FEMS Yeast Res. doi:10.1111/j.1567-1364.2009.00580.x (in press)
Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF (2005) Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22(5):359–368
la Grange DC, Pretorius IS, van Zyl WH (1996) Expression of a Trichoderma reesei beta-xylanase gene (XYN2) in Saccharomyces cerevisiae. Appl Environ Microbiol 62(3):1036–1044
Pronk JT (2002) Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol 68(5):2095–2100. doi:10.1128/aem.68.5.2095-2100.2002
Pronk JT, Steensma HY, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12(16):1607–1633
Rose MD, Winston F, Hieter P (1990) Methods in Yeast genetics: a cold spring harbor laboratory course manual. Cold Spring Harbor Laboratory Press, NY
Sherman F (2002) Getting started with yeast. Methods Enzymol 350:3–41
Shiba Y, Ono C, Fukui F, Watanabe I, Serizawa N, Gomi K, Yoshikawa H (2001) High-level secretory production of phospholipase A1 by Saccharomyces cerevisiae and Aspergillus oryzae. Biosci Biotechnol Biochem 65(1):94–101
Smith V, Botstein D, Brown PO (1995) Genetic footprinting: a genomic strategy for determining a gene’s function given its sequence. Proc Natl Acad Sci USA 92(14):6479–6483
Smith V, Chou KN, Lashkari D, Botstein D, Brown PO (1996) Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science 274(5295):2069–2074. doi:10.1126/science.274.5295.2069
van Dusen WJ, Fu J, Bailey FJ, Burke CJ, Herber WK, George HA (1997) Adenine quantitation in yeast extracts and fermentation media and its relationship to protein expression and cell growth in adenine auxotrophs of Saccharomyces cerevisiae. Biotechnol Prog 13(1):1–7
Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R (1994) Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem 269(13):9833–9841
Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11(1):53–55
Zhang J, Reddy J, Buckland B, Greasham R (2003) Toward consistent and productive complex media for industrial fermentations: Studies on yeast extract for a recombinant yeast fermentation process. Biotechnol Bioeng 82(6):640–652
Acknowledgments
This work was supported by Grants 3PR05A096 and PRI07A087 co-financed by the Junta de Extremadura and FEDER. I. Corbacho, F. Teixidó and R. Velázquez are recipients of pre-doctoral studentships from the Junta de Extremadura.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Corbacho, I., Teixidó, F., Velázquez, R. et al. Standard YPD, even supplemented with extra nutrients, does not always compensate growth defects of Saccharomyces cerevisiae auxotrophic strains. Antonie van Leeuwenhoek 99, 591–600 (2011). https://doi.org/10.1007/s10482-010-9530-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-010-9530-5