Abstract
The critical factors regulating stem cell endothelial commitment and renewal remain not well understood. Here, using loss- and gain-of-function assays together with bioinformatic analysis and multiple model systems, we show that PDGFD is an essential factor that switches on endothelial commitment of embryonic stem cells (ESCs). PDGFD genetic deletion or knockdown inhibits ESC differentiation into EC lineage and increases ESC self-renewal, and PDGFD overexpression activates ESC differentiation towards ECs. RNA sequencing reveals a critical requirement of PDGFD for the expression of vascular-differentiation related genes in ESCs. Importantly, PDGFD genetic deletion or knockdown increases ESC self-renewal and decreases blood vessel densities in both embryonic and neonatal mice and in teratomas. Mechanistically, we reveal that PDGFD fulfills this function via the MAPK/ERK pathway. Our findings provide new insight of PDGFD as a novel regulator of ESC fate determination, and suggest therapeutic implications of modulating PDGFD activity in stem cell therapy.
Similar content being viewed by others
Introduction
The platelet-derived growth factor (PDGF) family are known for their effects on mesenchyme and mural cells. Currently, there are four PDGFs (PDGFA, PDGFB, PDGFC, and PDGFD) and two receptors (PDGFR-α and PDGFR-β) [1]. PDGFD is a relatively new member of the family discovered many years after the finding of PDGFA and PDGFB, and binds to and activates PDGFR-β [1]. PDGFD has been shown to regulate fibrosis, inflammation, and tumorigenesis [2,3,4]. PDGFD has been reported to promote angiogenesis [5]. PDGFD is highly expressed in vascular cells in both mouse embryos and adults [6, 7], suggesting important roles of PDGFD in the vascular system. Pdgfd deficient mice are viable but display high blood pressure and disorganized pericytes in the cardiac vasculature [6], further suggesting a role of PDGFD in the regulation of the vascular system.
Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of blastocyst-stage embryos and have the capacity for self-renewal and pluripotency. Therefore, ESCs may have great therapeutic potential in regenerative medicine. Cardiovascular diseases, such as cardiac infarction, heart failure, and artery diseases, are the leading cause of mortality and morbidity worldwide [8]. Such diseases are often related to impaired vascular endothelial cell (EC) function. As such, derivation of functional and healthy ECs from ESCs has been a focus of interest in the field [9]. Recent advances in single cell RNA sequencing have provided new insight into stem cell-derived vascular cells [10]. Yet, critical molecules and mechanisms regulating ESC differentiation towards vascular cells remain poorly understood. Also, although PDGFD is expressed during embryonic development [7], it remains thus far unknown whether PDGFD plays a role in ESC regulation.
Here, we show that PDGFD is a critical factor that turns on ESC endothelial commitment. Pdgfd knockdown in ESCs increased ESC self-renewal and inhibited ESC differentiation into endothelial lineage in multiple assays. RNA sequencing analysis revealed that Pdgfd depletion caused transcriptome-wide downregulation of differentiation-related genes, particularly, vascular EC-related genes. Importantly, genetic deletion of Pdgfd in mice decreased cardiac blood vessel density in both embryonic and neonatal mice, and PDGFD knockdown in ESCs decreased blood vessel density during teratoma formation. Mechanistically, PDGFD fulfilled its function in ESCs via the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathway, and blocking MAPK/ERK signaling diminished PDGFD’s effect on ESCs. Collectively, our findings provide new insights into the mechanisms regulating ESC differentiation and vascular EC commitment, and suggest that modulating PDGFD activity may have therapeutic value in stem cell therapy.
Results
PDGFD is upregulated upon ESC differentiation
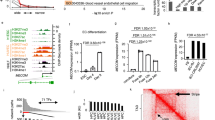
PDGFD is expressed during embryonic development [7]. Yet, it remains thus far unknown whether PDGFD plays a role in ESC regulation. We found that PDGFD expression in ESCs increased markedly upon ESC differentiation in multiple ESC differentiation assays, such as ESC differentiation induced by retinoic acid treatment (+RA), withdrawal of leukemia inhibitory factor (−LIF) and embryonic body (EB) formation. Both qRT-PCR (Fig. 1a, d, g) and Western blots (Fig. 1c, f, i) showed gradual increases of PDGFD transcript and protein levels during ESC differentiation with concomitant loss of SOX2, a key pluripotency factor (Fig. 1b, c, e, f, h, i), suggesting a potential effect of PDGFD on ESC differentiation. PDGFD is proteolytically cleaved by plasmin or uPA [11, 12], and cleaved forms of PDGFD were detected in ESC-conditioned serum-free medium with or without LIF by Western blot (Fig. S1a). Consistently, both plasmin and uPA were found during ESC differentiation under various conditions (Fig. S1b–d).
PDGFD is expressed in ESCs and upregulated upon ESC differentiation. a–c qRT-PCR (a, b) and Western blot (c) analyses showing PDGFD and SOX2 expression during ESC differentiation induced by retinoic acid treatment (+ RA). d–f qRT-PCR (d, e) and Western blot (f) analyses showing PDGFD and SOX2 expression during ESC differentiation induced by leukemia inhibitory factor withdrawal (−LIF). g–i qRT-PCR (g, h) and Western blot (i) analyses of PDGFD and SOX2 expression during ESC differentiation induced by embryoid body (EB) formation. In the experiments depicted in c, f, and i, densitometric quantification normalized to HSP90 and expressed relative to day 0 is shown beneath the blots. All data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using one-sample t-test in c, f, i and one-way ANOVA in a, b, d, e, g, h. *p < 0.05, **p < 0.01, ***p < 0.001
PDGFD promotes ESC differentiation and inhibits ESC self-renewal
To investigate the potential effect of PDGFD on ESCs, we knocked down Pdgfd (shPdgfd) in mouse ESCs, which was confirmed by qRT-PCR and Western blot (Fig. 2a, b). We found that PDGFD knockdown inhibited LIF withdrawal-induced ESC differentiation as shown by stronger AP staining, more compact colonies compared with control ESCs (shControl, Fig. 2c), decreased expression of many differentiation markers of the three germ layers and increased expression of Sox2, a key pluripotency marker (Fig. 2d–k). In addition, shPdgfd ESCs formed significantly smaller EBs (Fig. 2l, m) with downregulation of many differentiation markers, such as Gata4, Sox17, and Nestin (Fig. 2n–p) in an EB formation-induced ESC differentiation assay, demonstrating that PDGFD is critically required for ESC differentiation. In addition, in an EB formation assay, PDGFD knockdown upregulated the key ESC pluripotent markers Nanog, Oct4 and Sox2 as shown by qRT-PCR (Fig. 2q–s), suggesting increased ESC self-renewal. Indeed, ESCs with PDGFD knockdown formed more colonies in a colony formation assay (Fig. 2t, u). Moreover, the enhanced ESC self-renewal was further supported by the increased ESC proliferation as shown by the greater number of ESCs at different time points after plating (Fig. S2a). Together, these data demonstrate PDGFD induces ESC differentiation and inhibits ESC self-renewal.
PDGFD promotes ESC differentiation and inhibits ESC self-renewal. a, b qRT-PCR (a) and Western blot (b) showing PDGFD expression in ESCs with Pdgfd knockdown (shPdgfd). c Images showing shPdgfd ESCs cultured in the absence of LIF and after AP staining (lower panel). Scale bar 500 μm. d–k qRT-PCR (d–j) and Western blot (k) showing the expression of differentiation markers (d–i) and Sox2 (j, k) in shPdgfd ESCs. l, m Shown are EB formation of shPdgfd ESCs. EBs were imaged (l) and EB areas quantified (m). Scale bar 100 μm. AU arbitrary unit. n–p qRT-PCR analysis of the expression of differentiation markers in EBs derived from shPdgfd ESCs. q–s PDGFD knockdown upregulated the key ESC pluripotent markers Nanog, Oct4 and Sox2 in an EB formation assay as shown by qRT-PCR. t, u Results showing AP+ colonies of shControl or shPdgfd ESCs seven days after plating at different densities. In the experiments depicted in b and k, densitometric quantification normalized to HSP90 and expressed relative to shControl is shown beneath the blots. Data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using one-sample t-test in b, k and Student’s t-test in a, d–j, m–s, u. *p < 0.05, **p < 0.01, ***p < 0.001
To verify the specificity of the effect of PDGFD, we also investigated whether PDGFB had the same effect compared with PDGFD, since like PDGFD, PDGFB also binds to PDGFR-β [1]. By contrast to PDGFD, the knockdown of which increased ESC proliferation (Fig. S2a), Pdgfb knockdown slightly decreased ESC proliferation instead (Fig. S2b). Moreover, opposite to PDGFD, who’s knockdown up-regulated the pluripotency markers Nanog, Oct4 and Sox2 (Fig. 2q–s), Pdgfb knockdown decreased their expression instead (Fig. S2c–f), demonstrating that the effect of PDGFD was specific.
PDGFD is required for the expression of vascular-differentiation-related genes in ESCs
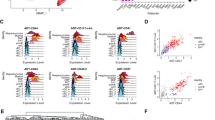
To investigate the genes regulated by PDGFD, we performed RNA sequencing using shPdgfd ESCs, and found 150 up- and 227 down-regulated genes in shPdgfd ESCs (> 1.5-fold change, p-value < 0.05, Fig. 3a). Gene Ontology (GO) analysis showed that the down-regulated genes in shPdgfd ESCs were mostly related to cell differentiation (Fig. 3b), such as Nsd2, Agpat1, Uso1, Elf4, Ncoa6, Mia, Gbx2, Dnmt3a, and Socs2 (Fig. 3c), and many of the up-regulated genes were linked to ESC pluripotency (Fig. S3a). The RNA sequencing results were confirmed by qRT-PCR (Fig. 3d–h). Importantly, GO term analysis revealed that many of the genes downregulated in shPdgfd ESCs were vascular-related, such as genes related to “angiogenesis”, “endothelium development’, “vasculature development”, “blood vessel morphogenesis” and “endothelial cell differentiation” (Fig. 3i), suggesting a role of PDGFD in these processes. In line with this, gene set enrichment analysis (GSEA) also unveiled that Pdgfd knockdown suppressed genes related to vascular endothelial-derived growth factor signaling pathways or tight junctions (Fig. 3j, k), both of which are essential for vascular development and function. These data thus demonstrate that PDGFD is critically required for the expression of vascular-differentiation-related genes in ESCs.
PDGFD is essential for the expression of vascular-differentiation-related genes in ESCs. a A volcano plot of RNA sequencing of shPdgfd ESCs showing 150 up- and 227 down-regulated genes (p < 0.05). b Gene Ontology (GO) analysis showing the top five GO terms of genes downregulated in shPdgfd ESCs. The dot size represents the number of differentially expressed genes of the GO term. c Top ten differentiation-related genes downregulated in shPdgfd ESCs. d–h qRT-PCR results showing the downregulation of the differentiation-related genes in shPdgfd ESCs. Data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using Student’s t-test. ***p < 0.001. i GO analysis showing the differentially expressed genes related to the vascular system in shPdgfd ESCs. j, k Gene set enrichment analysis (GSEA) depicting enrichment of VEGF signaling pathway (j) and tight junction (k)-related gene sets in shPdgfd ESCs. NES normalized enrichment score
PDGFD promotes endothelial cell (EC) commitment of ESCs
Led by our findings of the effect of PDGFD on the expression of vascular-related genes in ESCs, we subsequently investigated whether PDGFD plays a role in ESC differentiation into vascular ECs. In a VEGFA-induced EC differentiation assay, we found a marked upregulation of Pdgfd together with the EC markers Cdh5 and Kdr, with Pdgfd upregulation being earlier (Fig. 4a), suggesting a causative role of PDGFD in EC differentiation of ESCs. Consistently, in a VEGFA-induced EC differentiation assay, Pdgfd knockdown dampened the expression of the EC markers Cdh5 and Kdr (Fig. S3b, c). Moreover, PDGFD protein treatment of ESCs (Fig. 4b–e) or PDGFD overexpression (PDGFD OE, Fig. 4f–i) markedly increased the expression of EC markers, while Pdgfd knockdown decreased their expression (Fig. 4j, k). Different from PDGFD, who’s treatment up-regulated EC markers in ESCs, PDGFB treatment did not display such an effect (Fig. S2g, h). Noteworthy, these findings are consistent with previous reports showing different effects of PDGFD and PDGFB on adipose-derived stem cells [13] and support that the effects of PDGFD on ESCs were specific. Importantly, in a teratoma formation assay in vivo, Pdgfd knockdown decreased tumor blood vessel density and collagen IV expression (a marker for blood vessels) (Fig. 4l–n), with smaller tumor size and weight (Fig. 4o, p). Consistently, qRT-PCR showed decreased expression of EC markers (Cdh5 and Pecam1, Fig. 4q, r) and mesoderm markers (Hand1, Brachyury and Eomes) in shPdgfd-depleted teratomas (Fig. 4s–u). Collectively, these data demonstrate an important role of PDGFD in promoting EC commitment of ESCs and blood vessel formation.
PDGFD promotes ESC endothelial commitment. qRT-PCR results showing the expression of Pdgfd and endothelial markers (Cdh5 and Kdr) during endothelium differentiation of ESCs. b–e qRT-PCR (b–d) and Western blot (e) results showing that PDGFD protein treatment increased the expression of endothelial markers in ESCs (no VEGFA treatment). f–i Western blot (f) and qRT-PCR (g–i) results showing that overexpression of PDGFD (PDGFD OE) increased the expression of endothelial marker genes in ESCs (no VEGFA treatment). j, k qRT-PCR results showing decreased expression of endothelial marker genes in shPdgfd ESCs (no VEGFA treatment). l–n Immunohistochemistry staining for collagen IV was performed to identify blood vessels in the teratomas derived from shControl or shPdgfd ESCs (l). Quantification of collagen IV density and vessel numbers per field are shown in m and n. Scale bar 50 μm. o, p Shown are teratomas (o) and their weights (p) derived from shControl and shPdgfd ESCs. q–u qRT-PCR results showing the expression of endothelial cell (EC) markers (q, r) and mesoderm markers (s–u) of shControl and shPdgfd teratomas. Densitometric quantification normalized to GAPDH (e), HSP90 (f) and expressed relative to Control group is shown beneath the blots. Data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using one-sample t-test in e, f and one-way ANOVA in a and Student’s t-test in b–d, g–k, m, n, p–u. *p < 0.05, **p < 0.01, ***p < 0.001
MAPK/ERK signaling mediates the effects of PDGFD on ESCs
We next investigated the signaling pathway induced by PDGFD in ESCs. RNA-seq results showed that numerous genes of the MAPK/ERK pathways were downregulated by Pdgfd knockdown (Fig. 5a), indicating important functions of them in PDGFD-induced effects. Consistently, Western blot showed that Pdgfd knockdown reduced ERK phosphorylation (p-ERK) in ESCs, while STAT3, a known PDGFD downstream effector [5], was not affected (Fig. 5b). Moreover, PDGFD protein treatment activated ERK and PDGFR-β and up-regulated PDGFR-β at different time points in ESCs (Fig. 5c), consistent with previous reports that PDGFRs can be up-regulated rapidly within 10 min [14,15,16,17]. Furthermore, administration of PDGFR-β neutralizing antibody (nab) (Fig. 5d) or a PDGFR inhibitor Crenolanib (Fig. S4a) abolished PDGFD-induced ERK activation, indicating an important role of PDGFR-β in mediating PDGFD’s effects. Importantly, administration of an ERK inhibitor PD0325901 completely abolished PDGFD overexpression (PDGFD OE, Fig. S4b)-induced inhibition of ESC colony formation, AP staining (Fig. 5e), downregulation of Sox2, and upregulation of EC marker Cdh5 in ESCs (Fig. 5f, g). In addition, inhibition of ERK signaling by an ERK inhibitor PD0325901 up-regulated SOX2 (Fig. S4c) and down-regulated the EC markers Kdr and Pecam1 (Fig. S4d, e), thus phenocopying the effect of PDGFD knockdown. The ERK inhibitor PD0325901-induced SOX2 up-regulation was confirmed by Western blot (Fig. S4f).
MAPK/ERK signaling and PDGFR-β mediate effects of PDGFD on ESCs. a Heatmap showing the differentially expressed genes in shPdgfd ESCs related to MAPK/ERK signaling. b Western blot result showing reduced ERK phosphorylation in shPdgfd ESCs while STAT3 phosphorylation was not changed. c Western blot result showing PDGFD-induced ERK and PDGFR-β phosphorylation as well as PDGFR-β upregulation in ESCs. d Western blot result showing that administration of PDGFR-β neutralizing antibody (nab) abolished PDGFD-induced ERK phosphorylation in ESCs. e AP staining showing that administration of an ERK pathway inhibitor PD0325901 abolished PDGFD overexpression (PDGFD OE)-induced inhibition of colony formation in ESCs. Scale bar 300 μm. f, g qRT-PCR results showing that administration of an ERK pathway inhibitor PD0325901 abolished PDGFD overexpression (PDGFD OE)-induced down-regulation of Sox2 and up-regulation of Cdh5 in ESCs. Densitometric quantification normalized to HSP90 (b, d) and α-TUBULIN (c) and expressed relative to Control group is shown beneath the blots. Data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using one-way ANOVA in f, g and one-sample t-test in b–d. *p < 0.05, **p < 0.01, ***p < 0.001
It is known that SOX2 translocation from nucleus to cytoplasm promotes ESC differentiation [18]. We then tested whether ERK activation regulated this process by treating the ESCs with an ERK activator 12-O-tetradecanoylphorbol-13-acetate (TPA). We found rapid ERK phosphorylation (Fig. S4g) with concomitant translocation of SOX2 from nucleus to cytoplasm as shown by the reduced nucleus and increased cytoplasmic SOX2 levels (Fig. S4g). These data suggest that PDGFD-induced ESC differentiation may be fulfilled by ERK activation-induced SOX2 translocation from nucleus to cytoplasm.
Genetic deletion of Pdgfd impaired vascular endothelial cell development in mice
To verify the role of PDGFD in vivo, we generated Pdgfd-deficient mice (Fig. S5a–c). In embryonic day 3.5 (E3.5) embryos, from which ESCs are derived, Pdgfd deletion upregulated Sox2 and downregulated many differentiation marker genes (Fig. 6a–e), suggesting increased ESC renewal and decreased ESC differentiation. Indeed, Pdgfd knockout (KO) ESCs derived from E3.5 embryos formed more compact colonies with stronger AP staining (Fig. 6f), higher Sox2 expression levels and decreased expression of many EC markers, such as Kdr, Tie2, and Pecam1 (Fig. 6g–j), further showing increased ESC renewal. The vascular system forms between E10.5 and E13.5 during embryonic development [19]. Noteworthy, the expression of PDGFD and PDGFR-β increased during this time course together with the EC marker PECAM1 (Fig. 6k), suggesting important roles of PDGFD in vascular development. By contrast to PDGFD, the protein levels of PDGFB decreased during E10.5–E13.5 instead (Fig. S6a), suggesting a different function of PDGFB. Indeed, scRNA-seq analysis of E9.5–10.5 mouse embryos [20] also revealed markedly higher expression levels of PDGFD than those of PDGFB in endothelium (Fig. S6b), further supporting an important and unique role of PDGFD in endothelium formation. Importantly, in both E10.5 and E12.5 Pdgfd KO embryos, the expression of many vascular markers decreased (Fig. 6l–n). Noteworthy, PDGFD was mainly detected in the heart of mouse embryos at E12.5 (Fig. 7a) and was co-localized with PECAM1+ cells (Fig. 7b–d), demonstrating that PDGFD is highly expressed in the developing cardiac vasculature. In line with the reduced EC markers in Pdgfd−/− embryos, decreased blood vessel densities were found in the Pdgfd−/− hearts at both E12.5 (Fig. 7e, f) and postnatal day 1 (P1) (Fig. 7g, h) as revealed by PECAM1 staining. Together, these data demonstrate that genetic deletion of Pdgfd impaired vascular EC development in vivo.
Genetic deletion of Pdgfd impairs ESC differentiation in vivo and in vitro. a–e qRT-PCR result showing the upregulation of Sox2 and downregulation of differentiation-related genes in Pdgfd knockout (KO) E3.5 embryos. WT wild-type. f–j WT and Pdgfd KO ESCs were cultured without LIF and subjected to imaging (f, upper panel) and AP staining (f, lower panel). qRT-PCR results showing the upregulation of Sox2 and downregulation of EC markers in f (g–j). Scale bar 500 μm. k Western blot results showing the expression levels of PDGFD, PDGFR-β, PECAM1, and BRACHYURY in mouse embryos from E10.5 to E13.5. l–n qRT-PCR (l, m) and Western blot (n) results showing the expression levels of EC markers in WT and Pdgfd KO embryos. Densitometric quantification normalized to GAPDH (k, n) and expressed relative to E10.5 and WT group, respectively, is shown beneath the blots. Data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using one-sample t-test in k, n and Student’s t-test in a–e, g–j, l, m. *p < 0.05, **p < 0.01, ***p < 0.001
Loss of Pdgfd impairs vascular development in mice. a–d Immunofluorescence staining of PDGFD and PECAM1 in E12.5 mouse embryos. Shown in a is PDGFD staining on a section of a mouse embryo. Shown in b–d are high-magnification images of PDGFD and PECAM1 staining in the heart of an E12. 5 mouse embryo. Scale bars 20 μm. e, f Shown in e are PECAM1 immunofluorescence staining of hearts of E12.5 WT and Pdgfd KO embryos. Shown in f are the quantifications of e. Scale bars 100 μm on the top and 10 μm on the bottom. n = 4 for WT, and n = 5 for KO. g, h Shown in g are PECAM1 immunofluorescence staining of hearts of P1 WT and Pdgfd KO mice. Shown in h are the quantifications of g. Scale bars 200 μm on the left and 10 μm on the right. n = 5 for WT, and n = 6 for KO. Data are presented as mean ± SD, n = 3 each group. Statistical significance was determined using Student’s t-test in f and h. *p < 0.05, **p < 0.01
Discussion
In this study, we uncover an important role of PDGFD in promoting ESC endothelial differentiation and suppressing ESC self-renewal (Fig. 8). Genetic deletion of Pdgfd in mice impaired blood vessel formation in both embryonic development and teratoma growth, and overexpression of PDGFD-induced ESC differentiation towards EC lineage. Our findings suggest potential therapeutic implications of modulating PDGFD activity for stem cell therapy, particularly, for vascular diseases with EC defects.
PDGFD promotes ESC vascular differentiation and represses ESC self-renewal by activating ERK signaling via PDGFR-β. Higher levels of PDGFD expression in ESCs activates PDGFR-β and ERK, resulting in increased expression of EC markers and decreased SOX2 expression, thus increased ESCs differentiation into endothelial cells and decreased ESCs self-renewal
Traditionally, the PDGF family is known for their effects on mesenchymal cells, mural cells and tumor growth [1, 4]. Studies on PDGFs and their receptors in relationship to stem cells are limited with diverse observations. PDGFA has been shown to induce astrocyte differentiation in human ESCs [21], while another study showed that low concentrations of PDGFA maintain the undifferentiated state of ESCs [22]. PDGFB has been reported to promote ESC differentiation into smooth muscle cells [23] or cardiomyocytes [24], while it has also been shown that PDGF-BB, together with S1P, prevents ESC differentiation [25]. Observations on the effects of the two PDGF receptors on stem cells also differ. It was reported that PDGFR-α induces ESC differentiation into mesendoderm cells [26] and PDGFR-β promotes ESC differentiation into smooth muscle cells [27] or myeloid lineage [28], while it has also been shown that the PDGF receptors prevent ESC differentiation [22]. These findings suggest that the effects of the PDGFs and their receptors on ESCs might be context- or cell type-specific. Thus far, the potential effect of PDGFD on ESCs remains unclear.
Here, we found that during ESC differentiation, PDGFD expression increased by almost 10-folds, indicating an important role of PDGFD in this process. Indeed, loss of PDGFD by shRNA knockdown inhibited ESC differentiation and increased ESC self-renewal in multiple assays, such as colony formation, EB formation, and teratoma formation assays, demonstrating a key role of PDGFD in ESC differentiation. Moreover, Pdgfd knockdown in ESCs markedly decreased the protein level of BRACHYURY, a mesoderm marker, suggesting that PDGFD mainly affects ESC mesoderm commitment. In human mesenchymal stem cells (MSCs), PDGFD was shown to promote cell proliferation and sustain MSC multipotency [29]. The reasons for the different effects of PDGFD on ESCs and MSCs remain unclear thus far and future studies are required to address this.
Our RNA sequencing analysis showed that PDGFD-induced ESCs to differentiate towards vascular EC lineage. Indeed, Pdgfd knockdown decreased the expression of genes related to endothelium development, EC differentiation, and angiogenesis, and PDGFD treatment increased the expression of endothelial precursor cell (EPC) markers in ESCs. Consistently, and importantly, in vivo, decreased blood vessel densities were found in Pdgfd−/− (exon two of Pdgfd gene deleted) mouse embryos, neonatal mice and teratomas derived from shPdgfd ESCs. In a previously published work [6], the exon one of Pdgfd gene was replaced by a LacZ reporter gene and the Pdgfd−/− mice displayed a slightly higher blood pressure and disorganized NG2+ pericytes in the cardiac vasculature [6]. In addition, PDGFD was also found to be mainly expressed by vascular ECs [6], which is consistent with our results. However, this study did not investigate embryonic or neonatal Pdgfd−/− mice. Other studies have shown potent angiogenic effects of PDGFD [30, 31]. It would be important to know whether PDGFD-induced ECs could contribute to various physiological or pathological angiogenesis, and more studies are warranted to investigate into this.
The signaling pathway induced by PDGFD are not well understood thus far. In this work, RNA sequencing revealed that ERK signaling is a major pathway activated by PDGFD. PDGFD knockdown decreased ERK activation, and PDGFD protein treatment activated both ERK and PDGFR-β in ESCs. Furthermore, blocking the ERK pathway in ESCs completely abolished PDGFD-induced upregulation of EC markers, and increased SOX2 expression. ERK activation can promote ESC exit from pluripotency by regulating the phosphorylation of multiple transcription factors (TFs). For example, ERK activation phosphorylates NANOG and inhibits its transactivation by decreasing NANOG stability through ubiquitination [32]. Moreover, ERK activation initiates KLF4 nuclear export to the cytoplasm and leads to rapid ESC differentiation [33]. In ESCs, SOX2 harbors several putative ERK phosphorylation sites [34]. Yet, it remains unclear whether SOX2 activity or stability is regulated by ERK. In this study, we unveal that the nuclear fraction of SOX2 in ESCs decreased after ERK activation, thus raising the possibility that ERK might modulate SOX2 translocation in ESCs.
The observed effect of PDGFD on ESCs appeared to be specific, since PDGFB, the other PDGFRβ ligand, did not show the same effects in terms of ESC proliferation, inhibition of pluripotency genes, and EC marker induction. Indeed, these findings are consistent with previous reports showing different effects of PDGFD and PDGFB on adipose-derived stem cells [13]. We do not know the underlying mechanisms for the functional differences between PDGFD and PDGFB at this stage. Yet, multiple potential possibilities may exist. For example, the PDGFD and PDGFB genes use different enhancers. First, enhancer GH11J104162 regulates PDGFD but not PDGFB in ESCs and ECs (GeneCards). It is known that enhancer–promoter interactions can be stabilized and regulated by a group of proteins with similar functions [35]. The different protein-enhancer–promoter interactions might be involved in the functional aspects of PDGFD and PDGFB. Moreover, the PDGFD and PDGFB promoters have different TF binding sites (Table S1). Indeed, in mouse ESCs, ChIP-seq analysis (https://www.signalingpathways.org) revealed different TFs for Pdgfd and Pdgfb promoters (Table S2). It is known that different TFs may lead to distinct TF-dependent chromatin remodeling and gene expression [36, 37], and therefore different functional readouts. Further studies are needed to verify these.
In summary, our findings provide new insights into how EC commitment and renewal of ESCs are regulated by revealing PDGFD’s critical role in switching on ESC differentiation towards EC lineage while suppressing ESC renewal via the ERK pathway. Modulating PDGFD activity may have therapeutic potential in stem cell therapy for the treatment of various vascular diseases with EC defects.
Materials and methods
Mice
All mice were housed and maintained on a 12/12 h light/dark cycle at the Animal Facility of Zhongshan Ophthalmic Center, Sun Yat-Sen University. Six-week old nude mice were purchased from the GemPharmatech Company (Nanjing, China). Pdgfd deficient mice were generated by the Cyagen Company (Nanjing, China), and bred on a C57BL/6J background for more than six generations. Briefly, two gRNAs targeting Pdgfd exon 2 together with Cas9 mRNA were injected into mouse zygotes, which were subsequently transplanted into pseudopregnant mice. The neonatal mutant mice were genotyped by PCR and sequencing. The positive founder (F0) was bred to establish Pdgfd+/− mouse line. The primers used for genotyping the Pdgfd deficient mice by PCR are 5′-ATAACATAGTAAAGCGAAAACATGAACTG-3′ and 5′-GGCCACTCTTGTGGAAGATAATCTC-3′. A 928 bp PCR product represents the wild-type mice, and a 457 bp band represents the deletion of the Pdgfd gene. The absence of Pdgfd exon 2 mRNA in Pdgfd−/− mouse hearts (Fig. S5c) was confirmed by qRT-PCR. The primers are 5′-TTCCCGAACAGCTACCCAAG-3′ and 5′-TCCTTGGAGGGATCTCCTTGT-3′.
Cell culture and treatment
Mouse ESCs E14 (Darwin Core Facility, Baylor College of Medicine) were maintained in culture dish coated with 0.1% gelatin (Sigma-Aldrich, St. Louis, USA) in ESC medium containing KO DMEM medium (Thermo Fisher Scientific, Gibco, Waltham, MA, USA) and supplemented with 15% (v/v) fetal bovine serum (FBS; Gibco, Waltham, MA, USA), β-mercaptoethanol (0.1 µM), GlutaMax-I supplement (2 mM), MEM non-essential amino acids (0.1 mM), 1% (v/v) Penicillin–Streptomycin, LIF (1000 U/mL, Millipore, Billerica, MA, USA), and 2 inhibitors (CHIR99021, PD325901 at 3 µM and 1 µM, respectively) (Hejian Technology Co., Suzhou, China). HEK293T cells (COBIOER, Nanjing, China) were maintained in DMEM medium (Sigma-Aldrich, St. Louis, USA) with 10% (v/v) FBS, 1% Penicillin–Streptomycin. Mouse embryonic fibroblasts (MEFs, Cyagen, Santa Clara, CA, USA) serves as feeder cells and were maintained in DMEM medium with 10% FBS and 1% Penicillin–Streptomycin. The MEFs were treated with Mytomycin C (10 µg/mL, Sigma-Aldrich, St. Louis, USA) for 3 h before culturing in ESC medium. All cells were cultured at 37 °C in humidified air with 5% CO2.
For PDGFD or PDGFB protein stimulation, ESCs were starved in serum-free medium overnight and then stimulated with 50 ng/mL PDGFD protein (R&D, Minneapolis, MN, USA) or PDGFB protein (PeproTech, Cranbury, NJ, USA) for indicated timepoints. For ERK inhibition, the MEK inhibitor PD0325901 (1 μM) was applied for 2 days in ESC medium without 2i. For TPA treatment, cells were starved in serum-free medium overnight followed by treatment with 200 nM TPA (New England Biolabs, Ipswich, MA, USA) for 15 or 30 min. For Crenolanib treatment, cells were starved in serum-free medium overnight, followed by treatment with 50 ng/mL PDGFD protein together with 100 nM Crenolanib (Cayman Chemical, Ann Arbor, Michigan, USA) or control goat IgG (R&D, Minneapolis, MN, USA) for 30 min. The cells were then collected for Western blot.
ESC differentiation
For LIF withdrawal-induced ESC differentiation, ESCs were cultured in ESC medium in the absence of LIF for 3 days [38]. For RA-induced ESC differentiation, ESCs were cultured in ESCs medium containing 5 µM RA (Sigma-Aldrich, St. Louis, USA) for three days [39]. For embryoid body (EB)-induced ESC differentiation, EB formation was performed for 6 days (see detailed description below). For endothelium differentiation, 5 × 105 ESCs were cultured in a 100 mm petri dish in the absence of LIF for 2 days and cultured further with 30 ng/mL recombinant human VEGFA protein (PeproTech, Cranbury, NJ, USA) for 7 days [40].
Embryoid body (EB) formation
ESCs were seeded at a density of 4 × 104 cells/mL using a hanging drop (25 µL) method and cultured without LIF (−LIF) in ESC medium for six days in 150 mm dishes. At day 6, all the EBs were harvested, photographed and analyzed [41].
Detection of secreted PDGFD
Mouse ESCs were plated in two 10 cm dishes (1 × 106 each) and cultured in ESC medium. The next day, one dish was refreshed with normal ESC medium while the other switched to ESC medium without LIF (−LIF). After 48 h, ESCs were rinsed with PBS and maintained in 5 mL serum-free ESC medium with or without LIF for 2 days. The medium were then concentrated using Amicon Ultra-4 centrifugal filter units (UFC801024) and analyzed by Western blot. Ponceau S staining was used as a loading control.
PDGFB knockdown by siRNA
ESCs were transfected with siRNA oligos (GenePharma) targeting mouse PDGFB or scrambled siRNA as a control using Lipofectamine 2000 (Invitrogen). ESCs were trypsinized and 1 × 106 cells were seeded in a 60 mm culture dish with 4 mL medium/well. For each well, 200 pmol of siRNA was mixed with 500 µL of Opti-MEM (Thermo), and 10 µL Lipofectamine 2000 (Invitrogen) was mixed with another 500 µL of Opti-MEM, and incubated for 5 min. The two different mixtures were subsequently combined and incubated for 15 min, and added into each well. After 6 h, the culture medium was changed with new medium. Forty-eight hours after transfection, the ESCs were harvested for analysis. The sequences of the PDGFB siRNA oligos are: sense, 5′-GCCUGCAAGUGUGAGACAGUA-3′, and antisense, 5′-UACUGUCUCACACUUGCAGGC-3′.
PDGFR-β neutralization
ESCs were starved in serum-free medium overnight. The next day, ESCs were refreshed with serum-free medium and supplemented with 50 ng/mL PDGFD protein together with 0.6 μg/mL PDGFR-β receptor neutralizing antibody (Thermo Fisher Scientific, Invitrogen, Waltham, MA, USA) or control goat IgG (R&D, Minneapolis, MN, USA) for 30 min. The cells were then collected for Western blot analysis.
Generation of stable Pdgfd knockdown or overexpression ESCs
To generate stable Pdgfd knockdown ESCs, 1 μg of pLent-Pdgfd-4in1-shRNA plasmids (Vigene Biosciences, Rockville, MD, USA) expressing shRNAs targeting four different mouse Pdgfd sequences were transfected into HEK293T cells together with 2 μg of lentivirus packing plasmids pSPAX2 and pMD2G (Addgene, Watertown, MA, USA) using Lipofectamine 2000 (Life Technologies, Waltham, MA, USA). The supernatants containing the viral particles was collected after 48 h and added to the ESCs in the presence of 5 μg/mL of polybrene (Santa Cruz, Dallas, TX, USA). After 6 h, new ESC medium was added. The ESCs were selected using puromycin (2 μg/mL). The pLent-4in1-shRNA-GFP-Puro plasmid (Vigene Biosciences, Rockville, MD, USA) was used as a negative control. The stable pdgfd-overexpressing ESCs were constructed following the same procedure. Mouse pdgfd gene sequence was inserted into the plent-EF1a-FH-CMV-GFP-P2A-puro plasmid (Vigene Biosciences, Rockville, MD, USA). PCR primers used are 5′-CG GGATCCCGATGCAACGGCTCGTTTTAGT-3′ and 5′-ATAGCGGCCGCTCGAGGTGGTCTTGAGCTG CAGAT-3′. The shRNA sequences used are listed in Table S3.
Generation of Pdgfd −/− and Pdgfd +/+ ESCs
Pdgfd−/− and Pdgfd+/+ ESCs were derived from 3.5 day-old blastocyst-stage embryos obtained from Pdgfd+/− heterozygous breeding. Blastocysts were collected by flushing of oviducts and uteri using M2 media (Sigma-Aldrich, St. Louis, USA). Embryos were collected to four-well plates (Thermo Fisher Scientific, Waltham, MA, USA) with feeder cells in ESC medium. After 6–12 days, the ICM outgrowth was re-plated after trypsinization using 0.25% trypsin (Invitrogen Corp., Grand Island, NY, USA) on four-well plates with feeder cells. When the ESCs were sub-confluent, they were plated into larger gelatin-coated culture dishes. The ESCs were passaged every 2–4 days and genotyped by PCR.
Analysis of Pdgfd deficient embryos and neonatal mice
Pdgfd mutant embryos and neonatal mice were obtained from Pdgfd+/− × Pdgfd+/− breeding. Briefly, 6–8 weeks old female mice were superovulated by intraperitoneal injection of 5 IU pregnant mare’s serum gonadotropin (PMSG; Solarbio, Beijing, China), and after 48 h, followed by the injection of 5 IU human chorionic gonadotrophin (hCG; Millipore, Billerica, MA, USA). The female mice were then caged with male mice at a one-to-one ratio. The presence of a vaginal plug in a female mouse about 12–20 h after hCG injection marks 0.5 days post copulation.
For mouse blastocyst collection, embryos were flushed from oviducts and uteri using M2 media (Sigma-Aldrich, St. Louis, USA) at E3.5. The blastocysts were subjected to whole transcriptome amplification using the PEPLI-g WTA single cell kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The product was 10-fold diluted, and 1 µL of the diluted product was used for qRT-PCR.
For E10.5 or E12.5 embryo collection, the embryos were dissected from uterus, washed with phosphate buffered saline (PBS) and then grinded in RIPA (Solarbio, Beijing, China) or Trizol Reagent (Invitrogen, Waltham, MA, USA) and subjected to Western blot or qRT-PCR.
Colony formation assay and alkaline phosphatase (AP) staining
ESC colony formation assay was performed by plating the ESCs onto 0.1% gelatin-coated 6-well plates at different densities (200, 400, 800 cells per well) and culturing for seven days. Colony formation was analyzed by AP staining prior to imaging. AP staining was performed using the AP detection kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions.
Cell proliferation assay
To analyze ESC proliferation, 1 × 104 ESCs per well were plated in 12-well plates and cultured for 4 days. Cell numbers were quantified at 72 and 96 h using an automated cell counter (Inno-Alliance Biotech, Wilmington, DE, USA). Triplicate samples were used for each group at each time point.
RNA isolation, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using the Trizol reagent (Invitrogen, Waltham, MA, USA) following the manufacturer’s instructions. A total of 2 µg RNA was reverse transcribed to cDNA using the FastQuant RT Kit (Tiangen Biotech, Beijing, China), and then amplified with qRT-PCR via SYBR Green PCR Master Mix (Vazyme Biotech, Nanjing, China) using an ABI QuantStudio 6 Flex device (Life Technologies, Waltham, MA, USA). When the expressions of multiple genes were investigated, to show the overall expression levels, the expression of each gene was normalized against GAPDH using a ∆Ct method [∆Ct = Ct of the gene − Ct of GAPDH. Relative gene expression = \(2^{{ - (\Delta C_{{\text{t}}} )}}\)]. When the expression of only one gene was investigated, a ∆∆Ct method was used [∆∆Ct = ∆Ct of the gene − ∆Ct of control. Relative gene expression = \(2^{{ - (\Delta \Delta C_{{\text{t}}} )}}\)]. The sequences of the primers used are listed in Table S4.
Cellular fractionation
To obtain different cellular fractionations, ESCs were lysed using cytoskeletal buffer (50 mM NaCl, 300 mM sucrose, 10 mM Pipes, pH 6.8, 3 mM MgCl2, 0.5% Triton X-100, protease inhibitor (Beyotime Biotechnology, Shanghai, China), 1 mM DTT, 1 mM PMSF) for 30 min on ice. After centrifugation for 10 min at 4 °C, the cytoplasmic fraction in the supernatant was collected. The nuclear fraction in the pellet was washed, lysed in RIPA buffer (Solarbio, Beijing, China) and collected.
Western blot
Western blots were performed by separating proteins on SDS-PAGE gels followed by protein transfer to PVDF membranes (Bio-Rad). The antibodies used were: anti-PDGFD (sc-137030, Santa Cruz), anti-SOX2 (AF-2018, R&D), anti-BRACHYURY (sc-166962, Santa Cruz), anti-ERKs (4696, Cell Signaling Technology), anti-Phospho ERKs (4370, Cell Signaling Technology), anti-PDGFR-β (3169, Cell Signaling Technology), anti-Phospho PDGFR-β (Tyr1021) (2227, Cell Signaling Technology), anti-PECAM1 (222783, Abcom), anti-Phospho STAT3 (9145, Cell Signaling Technology), anti-STAT3 (9139, Cell Signaling Technology), anti-KDR (9698, Cell Signaling Technology), anti-HSP90 (7411, Cell Signaling Technology), anti-TUBULIN (RM2007, Ray Antibody), anti-β-ACTIN (RM2001, Ray Antibody), anti-GAPDH (70-Mab5465-040, MultiSciences), anti-HISTONE 3 (GB13102-1, Servicebio), Goat anti-mouse IgG (GAM0072, MultiSciences), Goat anti-rabbit IgG (GAR0072, MultiSciences), Rabbit anti-goat IgG (RAG0072, MultiSciences), anti-PDGFB (PA1-27394, Invitrogen), anti-uPA (17968-1-AP, Proteintech) and anti-PLASMIN (66399-1-Ig, Proteintech). The bands were visualized using a GBOX-CHEMI-XX8 device (SYNGENE).
Teratoma formation and immunohistochemistry analysis
The shControl or shPdgfd ESCs (1 × 106 cells/100 µL PBS) were injected subcutaneously into the dorsal flank regions of nude mice. Four weeks after injection, the teratomas were surgically dissected and weighed. Parts of the tumors were used for RNA extraction, and the rest were fixed in 4% paraformaldehyde, embedded in paraffin and sectioned (5 µm) for immunohistochemistry (IHC) staining.
For IHC staining, sections were placed at 60 °C for 1 h followed by deparaffinized with xylene and ethanol. Subsequently, the sections were processed in 10 mM citrate buffer (pH 6.0) and boiled for 10 min for antigen retrieval. After cooling in room temperature, the sections were incubated in 3% hydrogen peroxide for 10 min to block the endogenous peroxidase activity. The sections were stained with anti-Collagen IV (BioRad, Hercules, California, USA) overnight at 4 °C and then incubated using the Elivision kit (Maxim Biotech, Fuzhou, China) for 30 min at room temperature. The DAB color reagent (Maxim Biotech, Fuzhou, China) was used. Finally, hematoxylin staining was performed for nuclear visualization. Sections were visualized using a fluorescence microscope-Axio Imager Z2 (ZEISS, Oberkochen, Germany). ImageJ software was used to analyze the sections. Collagen IV density was calculated as Collagen IV+ pixels/total field pixels.
Immunofluorescence analysis
E12.5 mouse embryos or the hearts from P1 pups were fixed in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, USA) overnight at 4 °C, and then transferred into 30% sucrose overnight at 4 °C. The tissues were then embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Tokyo, Japan) and sectioned. The frozen sections were placed in room temperature for 30 min, blocked with 5% donkey serum, 0.5% Triton X-100 in 1 × PBS for one hour at room temperature. The sections were stained with anti-PECAM1 (BD Biosciences, New Jersey, USA) overnight at 4 °C and then stained with fluorescein-conjugated secondary antibodies (Life Technologies, Waltham, MA, USA) for 1 h at room temperature. Sections were visualized using a fluorescence microscope-Axio Imager Z2 (ZEISS, Oberkochen, Germany). All sections were imaged using a × 20 objective. The ImageJ software was used for image analysis. The ventricle areas of the hearts were imaged and analyzed. Vascular density was calculated as PECAM1+ pixels/total field pixels.
RNA sequencing (RNA-seq)
Total RNA was extracted from shControl or shPdgfd ESCs (triplicates for each group) using Trizol reagent (Invitrogen, Waltham, MA, USA). RNA-seq was performed using an Illumina Nova Seq 6000 sequenator (Illumina, San Diego, CA, USA) by Kangcheng Bio-tech, Inc. (Shanghai, China). The differentially expressed genes (DEGs) were defined at p < 0.05 and fold change > 1.5. The DEGs were subjected to GO analysis using the topGO package for enriched biological processes, and the KEGG pathway analysis using Fisher’s exact test for enriched pathways (p < 0.05). GSEA enrichment plots were generated using the GSEA software [42]. Default parameters were used for GSEA analysis. The RNA sequencing data were accessible in GEO database (GSE172117).
Statistical analysis
GraphPad Prism (GraphPad Software, Inc.) was used for statistical analyses except for the RNA-seq data. All data are presented as mean ± SD. For comparisons between two groups, two-tailed Student’s t tests were used. For comparison among more than two groups, one-way ANOVA analysis was used. N numbers, p values and other detailed information are provided in the corresponding figure legends.
Data availability
The data sets generated during the current study are available in the GEO database (GSE172117).
References
Papadopoulos N, Lennartsson J (2018) The PDGF/PDGFR pathway as a drug target. Mol Asp Med 62:75–88. https://doi.org/10.1016/j.mam.2017.11.007
Buhl E, Djudjaj S, Babickova J, Klinkhammer B, Folestad E, Borkham-Kamphorst E, Weiskirchen R, Hudkins K, Alpers C, Eriksson U, Floege J, Boor P (2016) The role of PDGF-D in healthy and fibrotic kidneys. Kidney Int 89(4):848–861. https://doi.org/10.1016/j.kint.2015.12.037
Yang P, Manaenko A, Xu F, Miao L, Wang G, Hu X, Guo Z, Hu Q, Hartman R, Pearce W, Obenaus A, Zhang J, Chen G, Tang J (2016) Role of PDGF-D and PDGFR-β in neuroinflammation in experimental ICH mice model. Exp Neurol 283:157–164. https://doi.org/10.1016/j.expneurol.2016.06.010
Jiang B, Chen J, Yuan W, Ji J, Liu Z, Wu L, Tang Q, Shu X (2018) Platelet-derived growth factor-D promotes colorectal cancer cell migration, invasion and proliferation by regulating Notch1 and matrix metalloproteinase-9. Oncol Lett 15(2):1573–1579. https://doi.org/10.3892/ol.2017.7510
Zhang J, Zhang H, Chen Y, Fu J, Lei Y, Sun J, Tang B (2019) Platelet-derived growth factor D promotes the angiogenic capacity of endothelial progenitor cells. Mol Med Rep 19(1):125–132. https://doi.org/10.3892/mmr.2018.9692
Gladh H, Folestad E, Muhl L, Ehnman M, Tannenberg P, Lawrence A, Betsholtz C, Eriksson U (2016) Mice lacking platelet-derived growth factor D display a mild vascular phenotype. PLoS ONE 11(3):e0152276. https://doi.org/10.1371/journal.pone.0152276
Pontén A, Folestad E, Pietras K, Eriksson U (2005) Platelet-derived growth factor D induces cardiac fibrosis and proliferation of vascular smooth muscle cells in heart-specific transgenic mice. Circ Res 97(10):1036–1045. https://doi.org/10.1161/01.RES.0000190590.31545.d4
Joseph P, Leong D, McKee M, Anand SS, Schwalm JD, Teo K, Mente A, Yusuf S (2017) Reducing the global burden of cardiovascular disease, Part 1: the epidemiology and risk factors. Circ Res 121(6):677–694. https://doi.org/10.1161/CIRCRESAHA.117.308903
Harding A, Cortez-Toledo E, Magner NL, Beegle JR, Coleal-Bergum DP, Hao D, Wang A, Nolta JA, Zhou P (2017) Highly efficient differentiation of endothelial cells from pluripotent stem cells requires the MAPK and the PI3K pathways. Stem Cells 35(4):909–919. https://doi.org/10.1002/stem.2577
Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, Rhee JW, Kim Y, Wirka RC, Buikema JW, Wu SM, Red-Horse K, Quertermous T, Wu JC (2018) Large-scale single-cell RNA-Seq reveals molecular signatures of heterogeneous populations of human induced pluripotent stem cell-derived endothelial cells. Circ Res 123(4):443–450. https://doi.org/10.1161/CIRCRESAHA.118.312913
Wang Z, Ahmad A, Li Y, Kong D, Azmi AS, Banerjee S (1806) Sarkar FH (2010) Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim Biophys Acta 1:122–130. https://doi.org/10.1016/j.bbcan.2010.04.003
Fredriksson L, Li H, Eriksson U (2004) The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev 15(4):197–204. https://doi.org/10.1016/j.cytogfr.2004.03.007
Kim WS, Park HS, Sung JH (2015) The pivotal role of PDGF and its receptor isoforms in adipose-derived stem cells. Histol Histopathol 30(7):793–799. https://doi.org/10.14670/HH-11-598
McGowan SE, McCoy DM (2018) Neuropilin-1 and platelet-derived growth factor receptors cooperatively regulate intermediate filaments and mesenchymal cell migration during alveolar septation. Am J Physiol Lung Cell Mol Physiol 315(1):L102–L115. https://doi.org/10.1152/ajplung.00511.2017
Shao W, Li X, Peng J, Fan S, Liang M, Huang K (2020) Apatinib attenuates phenotypic switching of arterial smooth muscle cells in vascular remodelling by targeting the PDGF Receptor-beta. J Cell Mol Med 24(17):10128–10139. https://doi.org/10.1111/jcmm.15623
Luk K, Boatman S, Johnson KN, Dudek OA, Ristau N, Vang D, Nguyen J, Gupta K (2012) Influence of morphine on pericyte–endothelial interaction: implications for antiangiogenic therapy. J Oncol 2012:458385. https://doi.org/10.1155/2012/458385
Huang W, Fridman Y, Bonfil R, Ustach C, Conley-LaComb M, Wiesner C, Saliganan A, Cher M, Kim H (2012) A novel function for platelet-derived growth factor D: induction of osteoclastic differentiation for intraosseous tumor growth. Oncogene 31(42):4527–4535. https://doi.org/10.1038/onc.2011.573
Baltus G, Kowalski M, Zhai H, Tutter A, Quinn D, Wall D, Kadam S (2009) Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells (Dayt Ohio) 27(9):2175–2184. https://doi.org/10.1002/stem.168
Walls J, Coultas L, Rossant J, Henkelman R (2008) Three-dimensional analysis of vascular development in the mouse embryo. PLoS ONE 3(8):e2853. https://doi.org/10.1371/journal.pone.0002853
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J (2019) The single-cell transcriptional landscape of mammalian organogenesis. Nature 566(7745):496–502. https://doi.org/10.1038/s41586-019-0969-x
Peng H, Chen G (2005) Neural precursors derived from human embryonic stem cells. Sci China C 48(3):295–299. https://doi.org/10.1360/062004-83
Brill L, Xiong W, Lee K, Ficarro S, Crain A, Xu Y, Terskikh A, Snyder E, Ding S (2009) Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5(2):204–213. https://doi.org/10.1016/j.stem.2009.06.002
Zhang L, Jin M, Margariti A, Wang G, Luo Z, Zampetaki A, Zeng L, Ye S, Zhu J, Xiao Q (2010) Sp1-dependent activation of HDAC7 is required for platelet-derived growth factor-BB-induced smooth muscle cell differentiation from stem cells. J Biol Chem 285(49):38463–38472. https://doi.org/10.1074/jbc.M110.153999
Sachinidis A, Gissel C, Nierhoff D, Hippler-Altenburg R, Sauer H, Wartenberg M, Hescheler J (2003) Identification of platelet-derived growth factor-BB as cardiogenesis-inducing factor in mouse embryonic stem cells under serum-free conditions. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol 13(6):423–429. https://doi.org/10.1159/000075130
Wong R, Pera M, Pébay A (2018) Maintenance of human embryonic stem cells by sphingosine-1-phosphate and platelet-derived growth factor. Methods Mol Biol (Clifton NJ) 1697:133–140. https://doi.org/10.1007/7651_2017_4
Pereira L, Wong M, Mossman A, Sourris K, Janes M, Knezevic K, Hirst C, Lim S, Pimanda J, Stanley E, Elefanty A (2012) Pdgfrα and Flk1 are direct target genes of Mixl1 in differentiating embryonic stem cells. Stem Cell Res 8(2):165–179. https://doi.org/10.1016/j.scr.2011.09.007
Shimizu N, Yamamoto K, Obi S, Kumagaya S, Masumura T, Shimano Y, Naruse K, Yamashita J, Igarashi T, Ando J (2008) Cyclic strain induces mouse embryonic stem cell differentiation into vascular smooth muscle cells by activating PDGF receptor beta. J Appl Physiol (Bethesda MD 1985) 104(3):766–772. https://doi.org/10.1152/japplphysiol.00870.2007
Dobbin E, Graham C, Corrigan P, Thomas K, Freeburn R, Wheadon H (2009) Tel/PDGFRbeta induces stem cell differentiation via the Ras/ERK and STAT5 signaling pathways. Exp Hematol 37(1):111–121. https://doi.org/10.1016/j.exphem.2008.09.012
Sotoca A, Roelofs-Hendriks J, Boeren S, van der Kraan P, Vervoort J, van Zoelen E, Piek E (2013) Comparative proteome approach demonstrates that platelet-derived growth factor C and D efficiently induce proliferation while maintaining multipotency of hMSCs. Exp Cell Res 319(17):2649–2662. https://doi.org/10.1016/j.yexcr.2013.07.027
Krenzlin H, Behera P, Lorenz V, Passaro C, Zdioruk M, Nowicki M, Grauwet K, Zhang H, Skubal M, Ito H, Zane R, Gutknecht M, Griessl M, Ricklefs F, Ding L, Peled S, Rooj A, James C, Cobbs C, Cook C, Chiocca E, Lawler S (2019) Cytomegalovirus promotes murine glioblastoma growth via pericyte recruitment and angiogenesis. J Clin Investig 129(4):1671–1683. https://doi.org/10.1172/jci123375
Kumar A, Hou X, Lee C, Li Y, Maminishkis A, Tang Z, Zhang F, Langer H, Arjunan P, Dong L, Wu Z, Zhu L, Wang L, Min W, Colosi P, Chavakis T, Li X (2010) Platelet-derived growth factor-DD targeting arrests pathological angiogenesis by modulating glycogen synthase kinase-3beta phosphorylation. J Biol Chem 285(20):15500–15510. https://doi.org/10.1074/jbc.M110.113787
Kim S, Kim M, Cho Y, Yao K, Kim D, Jeong C, Yu D, Bae K, Cho E, Jung S, Lee M, Chen H, Kim J, Bode A, Dong Z (2014) ERK1 phosphorylates Nanog to regulate protein stability and stem cell self-renewal. Stem Cell Res 13(1):1–11. https://doi.org/10.1016/j.scr.2014.04.001
Dhaliwal N, Miri K, Davidson S, Tamim El Jarkass H, Mitchell J (2018) KLF4 nuclear export requires ERK activation and initiates exit from naive pluripotency. Stem Cell Rep 10(4):1308–1323. https://doi.org/10.1016/j.stemcr.2018.02.007
Brumbaugh J, Hou Z, Russell J, Howden S, Yu P, Ledvina A, Coon J, Thomson J (2012) Phosphorylation regulates human OCT4. Proc Natl Acad Sci USA 109(19):7162–7168. https://doi.org/10.1073/pnas.1203874109
Mora A, Sandve GK, Gabrielsen OS, Eskeland R (2016) In the loop: promoter–enhancer interactions and bioinformatics. Brief Bioinform 17(6):980–995. https://doi.org/10.1093/bib/bbv097
Lin J, Zhao Y, Ferraro AR, Yang E, Lewis ZA, Lin X (2019) Transcription factor Znf2 coordinates with the chromatin remodeling SWI/SNF complex to regulate cryptococcal cellular differentiation. Commun Biol 2:412. https://doi.org/10.1038/s42003-019-0665-2
Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M (2011) Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23(6):2285–2301. https://doi.org/10.1105/tpc.111.085266
Cherepkova M, Sineva G, Pospelov V (2016) Leukemia inhibitory factor (LIF) withdrawal activates mTOR signaling pathway in mouse embryonic stem cells through the MEK/ERK/TSC2 pathway. Cell Death Dis 7:e2050. https://doi.org/10.1038/cddis.2015.387
Tighe A, Gudas L (2004) Retinoic acid inhibits leukemia inhibitory factor signaling pathways in mouse embryonic stem cells. J Cell Physiol 198(2):223–229. https://doi.org/10.1002/jcp.10424
Boeckel J, Derlet A, Glaser S, Luczak A, Lucas T, Heumüller A, Krüger M, Zehendner C, Kaluza D, Doddaballapur A, Ohtani K, Treguer K, Dimmeler S (2016) JMJD8 regulates angiogenic sprouting and cellular metabolism by interacting with pyruvate kinase M2 in endothelial cells. Arterioscler Thromb Vasc Biol 36(7):1425–1433. https://doi.org/10.1161/atvbaha.116.307695
Keller G (1995) In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol 7(6):862–869. https://doi.org/10.1016/0955-0674(95)80071-9
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Acknowledgements
This study was supported by the State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-Sen University, Guangzhou, China; Program of Guangzhou Scientific Research Plan (202102010179) to X.L., Natural Science Foundation of Guangdong Province grant (2019A1515010229, 2021A1515010684) to W.L.; the Guangdong Provincial Key Laboratory of Ophthalmology and Visual Science (2020B1212060033) to W.L., Guangzhou Basic and Applied Research Program (202102020146) to W.L., and National Natural Science Foundation of China (82150710555 to X.L.; 82070983 and 81870679 to Q.J).
Author information
Authors and Affiliations
Contributions
WL and XL designed and supervised all experiments. PX, BD, YZ, XL, JZ, XX, ZX, XY, XZ, LH, BX, CW, SW, HK, XH, WL, and AM performed experiments and analyzed data. WL, XL, YC and QJ supervised work and conceptualized data. WL, XL, and PX wrote the paper. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All animal experiments were approved by the Animal Care and Use Committee at the Zhongshan Ophthalmic Center, Sun Yat-Sen University, People’s Republic of China (Protocol Number 2020-170).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, W., Xu, P., Deng, B. et al. PDGFD switches on stem cell endothelial commitment. Angiogenesis 25, 517–533 (2022). https://doi.org/10.1007/s10456-022-09847-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10456-022-09847-4