Abstract
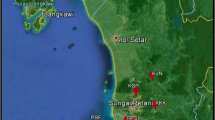
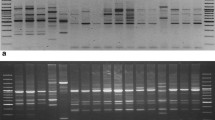
In this study, genetic diversity and phylogenetic analysis of Turkish Juglans regia L. populations was conducted based on random-amplified polymorphic DNA (RAPD) and inter-simple sequence repeat polymerase chain reaction ISSR-PCR and nuclear ribosomal DNA internal transcribed spacer (nrDNA ITS) sequences. A total of 14 populations from the Aegean and Marmara regions were included in the research. In all, 12 RAPD and 13 ISSR primers were used for molecular characterization of the J. regia populations. For the amplification of the nrDNA ITS region, ITS4 and ITS5A primers were used in PCR. In RAPD-ISSR-PCR analysis, a total of 55 and 85 bands were obtained, respectively. RAPD and ISSR analyses detected high polymorphism (74.54% and 70.57%, respectively) among J. regia populations. However, ITS sequence analysis was not efficient in revealing the genetic relationship among J. regia samples. In addition, phylogenetic analyses including nrDNA ITS sequences of our populations and that of other Juglans taxa (J. hindsii, J. major, J. nigra, J. microcarpa, J. cathayensis and J. hopeiensis) obtained from the National Center for Biotechnology Information (NCBI) were conducted. Analysis of J. regia populations based on RAPD and ISSR-PCR and ITS sequences revealed that the rate of polymorphism obtained with the ISSR-PCR approach was higher than those obtained using the ITS sequences and RAPD-PCR. Genetic diversity results of these populations will contribute significantly to the identification and breeding programs of walnuts.





Similar content being viewed by others
References
Al-Hemaid FM, Ali MA, Lee J, Kim SY, Rahman MO (2015) Molecular evolutionary relationships of Euphorbia scordifolia Jacq. within the genus inferred from analysis of internal transcribed spacer sequences. Bangladesh J Plant Taxon 22(2):111–118. https://doi.org/10.3329/bjpt.v22i2.26072
Arab MM, Marrano A, Abdollahi-Arpanahi R, Leslie CA, Askari H, Neale DB, Vahdati K (2019) Genome-wide patterns of population structure and association mapping of nut-related traits in Persian walnut populations from Iran using the Axiom J. regia 700K SNP array. Sci Rep 9(1):1–14. https://doi.org/10.1038/s41598-019-42940-1
Aradhya MK, Potter D, Gao F, Simon CJ (2007) Molecular phylogeny of Juglans (Juglandaceae): a biogeographic perspective. Tree Genet Genomes 3(4):363–378. https://doi.org/10.1007/s11295-006-0078-5
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277. https://doi.org/10.2307/2399880
Boruzi AI, Nour V (2019) Walnut (Juglans regia L.) leaf powder as a natural antioxidant in cooked pork patties. Cyta J Food 17(1):431–438. https://doi.org/10.1080/19476337.2019.1596984
Çifçi EA, Yağdı K (2012) Study of genetic diversity in wheat (Triticum aestıvum) varities using random amplified polymorphic DNA (RAPD) analysis. Turk J Field Crop 17(1):91–95
Cosmulescu S, Trandafir I, Achim G, Mihai B, Baciu A, Gruia M (2010) Phenolics of green husk in mature walnut fruits. Not Bot Hortic Agrobot 38(1):53–56. https://doi.org/10.15835/nbha3814624
Costa R, Pereira G, Garrido I, Tavares-de-Sousa MM, Espinosa F (2016) Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify orchardgrass (Dactylis glomerata L.) germplasm variations. PLoS ONE 11(4):e152972. https://doi.org/10.1371/journal.pone.0152972
Cubero OF, Crespo A, Esslinger TL, Lumbsch HT (2004) Molecular phylogeny of the genus Physconia (Ascomycota, Lecanorales) inferred from a Bayesian analysis of nuclear ITS rDNA sequences. Mycol Res 108(5):498–505. https://doi.org/10.1017/s095375620400975x
Dhakshanamoorthy D, Selvaraj R, Chidambaram A (2015) Utility of RAPD marker for genetic diversity analysis in gamma rays and ethyl methane sulphonate (EMS)-treated Jatropha curcas plants. CR Biol 338(2):75–82. https://doi.org/10.1016/j.crvi.2014.12.002
Dolarslan M, Gül E, Acar E, Turktas M (2017) A phylogenetic perspective on the influence of ecological attributes on selected species of Astragalus. Ecoscience 24(3–4):105–113. https://doi.org/10.1080/11956860.2017.1373424
Dönmez IE, Güler HK (2015) Chemical composition of walnut shell. Res J Biol Sci 8:24–26
Ebrahimi A, Zarei A, Fardadonbeh MZ, Lawson S (2017) Evaluation of genetic variability among “Early Mature” Juglans regia using microsatellite markers and morphological traits. PeerJ 5:e3834. https://doi.org/10.7717/peerj.3834
Ercisli S, Orhan E, Esitken A, Yildirim N, Agar G (2008) Relationships among some cornelian cherry genotypes (Cornus mas L.) based on RAPD analysis. Genet Resour Crop Evol 55(4):613–618. https://doi.org/10.1007/s10722-007-9266-x
Ercisli S, Ipek A, Barut E (2011) SSR marker-based DNA fingerprinting and cultivar identification of olives (Olea europaea). Biochem Genet 49(9):555–561. https://doi.org/10.1007/s10528-011-9430-z
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Noori A, Mahdavian Z, Moradi Z (2016) Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip 30(6):1075–1081. https://doi.org/10.1080/13102818.2016.1228478
Feliner GN, Rosselló JA (2007) Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol Phylogenet Evol 44(2):911–919. https://doi.org/10.1016/j.ympev.2007.01.013
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamouda M (2019) Molecular analysis of genetic diversity in population of Silybum marianum (L.) Gaertn in Egypt. J Genet Eng Biotechnol 17(1):1–9
He HY, Zhang D, Qing H, Yang Y (2016) Analysis of the genetic diversity of Lonicera japonica Thumb. using inter-simple sequence repeat markers. Genet Mol Res 16:1–7. https://doi.org/10.4238/gmr16019338
Hocaoglu-Ozyigit A, Ucar B, Altay V, Ozyigit II (2022) Genetic diversity and phylogenetic analyses of Turkish cotton (Gossypium hirsutum L.) lines using ISSR markers and chloroplast trnL‑F regions. J Nat Fibers 19(5):1837–1850. https://doi.org/10.1080/15440478.2020.1788493
Hürkan K (2019) Anadolu Orkidesi’nde (Orchis anatolica Boiss.) Ribozomal DNA’nın Transkripsiyonu Yapılan İç Ara Bölgelerinin (ITS) Özellikleri. Igdır Univ Fen Bilim Enstitüsü Derg 9(2):1117–1127. https://doi.org/10.21597/jist.489021
Ipek M, Arıkan S, Pırlak L, Esitken A (2019) Phenological, morphological and molecular characterization of some promising walnut (Juglans regia L) genotypes in Konya. Erwerbs-Obstbau 61:149–156. https://doi.org/10.1007/s10341-018-0411-9
Kabiri G, Bouda S, Elhansali M, Haddioui A (2019) Genetic diversity and structure of walnut (Juglans regia L.) genotypes from Middle and High Atlas mountains of Morocco as investigated by Inter-Simple Sequence Repeat (ISSR) markers. Aust J Crop Sci 13(12):1983–1991
Kafkas S, Ozkan H, Sutyemez M (2005) DNA polymorphism and assessment of genetic relationships in walnut genotypes based on AFLP and SAMPL markers. J Am Soc Hortic Sci 130(4):585–590. https://doi.org/10.21273/JASHS.130.4.585
Kaya I, Demir I, Usta M, Sipahioğlu HM (2020) Molecular phylogeny based on its sequences of nrDNA of some species belonging to dodder (Cuscuta L.) genus from various ecological sites of Turkey. Notulae Bot Horti Agrobot Cluj Napoca 48(3):1332–1340. https://doi.org/10.15835/nbha48311997
Kiani G, Siahchehreh M (2018) Genetic diversity in tomato varieties assessed by ISSR markers. Int J Veg Sci 24(4):353–360. https://doi.org/10.1080/19315260.2017.1419397
Koloren O, Koloren Z, Eker S (2016) Molecular phylogeny of Artemisia species based on the internal transcribed spacer (ITS) of 18S-26S rDNA in Ordu Province of Turkey. Biotechnol Biotechnol Equip 30(5):929–934. https://doi.org/10.1080/13102818.2016.1188674
Korkmaz M, Dogan NY (2018) Analysis of genetic relationships between wild roses (Rosa L. Spp.) growing in Turkey. Erwerbs-Obstbau 60(4):305–310. https://doi.org/10.1007/s10341-018-0375-9
Liu K, Zhao S, Wang S, Wang H, Zhang Z (2020) Identification and analysis of the FAD gene family in walnuts (Juglans regia L.) based on transcriptome data. BMC Genomics 21:299. https://doi.org/10.1186/s12864-020-6692-z
Liu R, Wu L, Du Q, Ren JW, Chen QH, Li D, Mao R‑X, Liu X‑R, Li Y (2019) Small molecule oligopeptides isolated from walnut (Juglans regia L.) and their anti-fatigue effects in mice. Molecules 24(1):45. https://doi.org/10.3390/molecules24010045
Ma QG, Zhang JP, Pei D (2020) Molecular genetic variability of Juglans regia L. and Juglans sigillata D. as revealed by fluorescent amplified-fragment length polymorphism. Biotechnol Biotechnol Equip 34(1):725–731. https://doi.org/10.1080/13102818.2020.1805016
Malik SK, Rohini MR, Kumar S, Choudhary R, Pal D, Chaudhury R (2012) Assessment of genetic diversity in Sweet Orange [Citrus sinensis (L.) Osbeck] cultivars of India using morphological and RAPD markers. Agric Res 1(4):317–324
Maras M (2008) An ITS DNA sequence-based phylogenic study of some Heracleum L.(Umbelliferae) species from Turkey’s partial flora. Turk J Biochem 33(4):163–168
Maraş-Vanlıoğlu FG, Yaman H, Kayaçetin F (2020) Genetic diversity analysis of some species in Brassicaceae family with ISSR markers. Biotech Stud 29(1):38–46. https://doi.org/10.38042/biost.2020.29.01.05
Martín MP, LaGreca S, Lumbsch HT (2003) Molecular phylogeny of Diploschistes inferred from ITS sequence data. Lichenologist 35(1):27–32. https://doi.org/10.1006/lich.2002.0427
Mei Z, Zhang X, Khan MA, Imani S, Liu X, Zou H, Wei C, Fu J (2017) Genetic analysis of Penthorum chinense Pursh by improved RAPD and ISSR in China. Electron J Biotechnol 30:6–11. https://doi.org/10.1016/j.ejbt.2017.08.008
Mostajeran F, Yousefzadeh H, Davitashvili N, Kozlowski G, Akbarinia M (2017) Phylogenetic relationships of Pterocarya (Juglandaceae) with an emphasis on the taxonomic status of Iranian populations using ITS and trnH-psbA sequence data. Plant Biosystems 151(6):1012–1021. https://doi.org/10.1080/11263504.2016.1219416
Mu XY, Tong L, Sun M, Zhu YX, Wen J, Lin QW, Liu B (2020) Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol Phylogenet Evol 147:106802. https://doi.org/10.1016/j.ympev.2020.106802
Negi AS, Luqman S, Srivastava S, Krishna V, Gupta N, Darokar MP (2011) Antiproliferative and antioxidant activities of Juglans regia fruit extracts. Pharm Biol 49(6):669–673. https://doi.org/10.3109/13880209.2010.537666
Nicese FP, Hormaza JI, McGranahan GH (1998) Molecular characterization and genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers. Euphytica 101:199–206. https://doi.org/10.1023/A:1018390120142
Orhan E, Eyduran SP, Poljuha D, Akin M, Weber T, Ercisli S (2020) Genetic diversity detection of seed-propagated walnut (Juglans regia L.) germplasm from Eastern Anatolia using SSR markers. Folia Hortic 32(1):37–46
Ousmael KM, Tesfaye K, Hailesilassie T (2019) Genetic diversity assessment of yams (Dioscorea spp.) from Ethiopia using inter simple sequence repeat (ISSR) markers. Afr J Biotechnol 18(30):970–977. https://doi.org/10.5897/AJB2018.16446
Potter D, Gao F, Aiello G, Leslie C, McGranahan G (2002) Intersimple sequence repeat markers for fingerprinting and determining genetic relationships of walnut (Juglans regia) cultivars. J Am Soc Hortic Sci 127(1):75–81. https://doi.org/10.21273/JASHS.127.1.75
Rather MA, Dar BA, Dar MY, Wani BA, Shah WA, Bhat BA, Ganai BA, Bhat KA, Anand R, Qurishi MA (2012) Chemical composition, antioxidant and antibacterial activities of the leaf essential oil of Juglans regia L. and its constituents. Phytomedicine 19(13):1185–1190. https://doi.org/10.1016/j.phymed.2012.07.018
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Salieh FMH, Tahir NAR, Faraj JM (2013) Assessment of genetic relationship among some Iraqi walnut genotypes (Juglans regia L.) in Sulaimani region using RAPD and SSR molecular markers. Jordan J Agric Sci 9(3):351–362
Soilhi Z, Trindade H, Vicente S, Gouiaa S, Khoudi H, Mekki M (2020) Assessment of the genetic diversity and relationships of a collection of Mentha spp. in Tunisia using morphological traits and ISSR markers. J Hortic Sci Biotechnol 95(4):483–495. https://doi.org/10.1080/14620316.2019.1702482
Son JH, Park KC, Lee SI, Kim JH, Kim NS (2012) Species relationships among Allium species by ISSR analysis. Hortic Environ Biotechnol 53:256–262. https://doi.org/10.1007/s13580-012-0130-3
Stanford AM, Harden R, Parks CR (2000) Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS sequence data. Am J Bot 87(6):872–882. https://doi.org/10.2307/2656895
Sultana S, Lim YP, Bang JW, Choi HW (2011) Internal transcribed spacer (ITS) and genetic variations in Lilium native to Korea. Hortic Environ Biotechnol 52:502–510. https://doi.org/10.1007/s13580-011-0050-7
Sun YL, Park WG, Oh HK, Hong SK (2014) Genetic diversity and hybridization of Pulsatilla tongkangensis based on the nrDNA ITS region sequence. Biologia 69(1):24–31. https://doi.org/10.2478/s11756-013-0284-1
Sütyemez M, Güvenç G, Bükücü ŞB, Özcan A (2019) The determination of genetic diversity among some sumac (Rhus coriaria L.) genotypes. Erwerbs-Obstbau 61(4):355–361. https://doi.org/10.1007/s10341-019-00440-6
Swofford DL (2001) PAUP phylogenetic analysis using parsimony (and other methods), version 4.0b10 for 32-bit Microsoft Windows. Sinauer Associates, Sunderland
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tiwari M, Singh NK, Rathore M, Kumar N (2005) RAPD markers in the analysis of genetic diversity among common bean germplasm from Central Himalaya. Genet Resour Crop Evol 52(3):315–324. https://doi.org/10.1007/s10722-005-5123-y
Wang H, Pan G, Ma Q, Zhang J, Pei D (2015) The genetic diversity and introgression of Juglans regia and Juglans sigillata in Tibet as revealed by SSR markers. Tree Genet Genomes 11:804. https://doi.org/10.1007/s11295-014-0804-3
White TJ, Bruns T, Lee S, Taylor J (1990) Amplifications and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand J, Sninsky T (eds) PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, CA, USA, pp. 315–322
Yang S, Li X, Huang F, Huang Y, Liu X, Wu J, Wang Q, Deng Z, Chen R, Zhang M (2018) A new method based on SNP of nrDNA-ITS to identify Saccharum spontaneum and its progeny in the genus Saccharum. PLoS ONE 13(5):e197458. https://doi.org/10.1371/journal.pone.0197458
Yerlikaya C, Yucel S, Erturk Ü, Korukluoğlu M (2012) Proximate composition, minerals and fatty acid composition of Juglans regia L. genotypes and cultivars grown in Turkey. Braz Arch Biol Technol 55(5):677–683. https://doi.org/10.1590/S1516-89132012000500006
Yigit T, Saritepe Y, Özer AS, Aslan A, Erdogan A (2013) Determination of some physical and chemical properties of walnuts (Juglans regia L.) selected from Hekimhan (Malatya) region. Fruit Sci 1(1):41–45
Yıldırım E, Sütyemez M (2020) Sütyemez‑1 Ceviz Çeşidinden Açık Tozlanma ile Elde Edilen F1 Bitkilerinin Fenolojik ve Moleküler Karakterizasyonu. Yüzüncü Yıl Univ Tarım Bilim Derg 30(2):299–309. https://doi.org/10.29133/yyutbd.706034
Zarini HN, Jafari H, Ramandi HD, Bolandi AR, Karimishahri MR (2019) A comparative assessment of DNA fingerprinting assays of ISSR and RAPD markers for molecular diversity of Saffron and other Crocus spp. in Iran. Nucleus 62(1):39–50. https://doi.org/10.1007/s13237-018-0261-8
Funding
This research was supported by the TUBITAK 2017/2209- A (Project no: 1919B011702124).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
E. Sevindik, K. Okan, M. Sevindik and S. Ercisli declare that they have no competing interests.
Rights and permissions
Springer Nature oder sein Lizenzgeber (z.B. eine Gesellschaft oder ein*e andere*r Vertragspartner*in) hält die ausschließlichen Nutzungsrechte an diesem Artikel kraft eines Verlagsvertrags mit dem/den Autor*in(nen) oder anderen Rechteinhaber*in(nen); die Selbstarchivierung der akzeptierten Manuskriptversion dieses Artikels durch Autor*in(nen) unterliegt ausschließlich den Bedingungen dieses Verlagsvertrags und dem geltenden Recht.
About this article
Cite this article
Sevindik, E., Okan, K., Sevindik, M. et al. Genetic Diversity and Phylogenetic Analyses of Juglans regia L. (Juglandaceae) Populations Using RAPD, ISSR Markers and nrDNA ITS Regions. Erwerbs-Obstbau 65, 311–320 (2023). https://doi.org/10.1007/s10341-023-00834-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10341-023-00834-7