Abstract
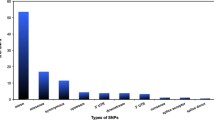
The ApoE gene responsible for the Alzheimer's disease has been examined to identify functional consequences of single-nucleotide polymorphisms (SNPs). Eighty-eight SNPs have been identified in the ApoE gene in which 31 are found to be nonsynonymous, 8 of them are coding synonymous, 33 are found to be in intron, and 3 are in untranslated region. The SNPs found in the untranslated region consisted of two SNPs from 5′ and one SNP from the 3′. Twenty-nine percent of the identified nsSNPs have been reported as damaging. In the analysis of SNPs in the UTR regions, it has been recognized that rs72654467 from 5′ and rs71673244 from 5′ and 3′ are responsible for the alteration in levels of expression. Both native and mutant protein structures were analyzed along with the stabilization residues. It has been concluded that among all SNPs of ApoE, the mutation in rs11542041 (R132S) has the most significant effect on functional variation.
Similar content being viewed by others
References
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Artiga MJ, Bullido MJ, Frank A et al (1998) Risk for Alzheimer's disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet 7:1887–1892
Bekris LM, Millard SP et al (2008) Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis 13:255–266
Chasman D, Adams RM (2001) Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J Mol Biol 307:683–706
Corder EH et al (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923
Emahazion T, Feuk L, Jobs M et al (2001) SNP association studies in Alzheimer's disease highlight problems for complex disease analysis. Trends Genet 17:407–413
Grillo G, Turi A et al (2010) UTRdb and UTRsite (RELEASE 2010)—a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38:75–80
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer—an environment for comparative protein modelling. Electrophoresis 18:2714–2723
Laws SM, Hone E, Gandy S, Martins RN (2003) Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem 84:1215–1236
Magyar C, Michael Gromiha M, Pujadas G, Tusnády GE, Simon I (2005) SRide—a server for identifying stabilizing residues in proteins. Nucleic Acids Res 33:303–305
Michikawa M (2003) The role of cholesterol in pathogenesis of Alzheimer's disease—dual metabolic interaction between amyloid β-protein and cholesterol. Mol Neurol 27:1–12
Ng PC, Henikoff S (2002) Accounting for human polymorphisms predicted to affect protein function. Genome Res 12:436–446
Pauline C, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11:863–874
Pesole G, Liuni S (1999) Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Trends Genet 15:378
Prokunina L, Alarcon-Riquelme ME (2004) Regulatory SNPs in complex diseases: their identification and functional validation. Expert Rev Mol Med 6:10
Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A (2011) Critical association of ncRNA with introns. Nucleic Acids Res 39:2357–2366
Ryan M, Diekhans M, Lien S, Liu Y, Karchin R (2009) LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics 25:1431–1432
Schork NJ, Fallin D, Lanchbury JS (2000) Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet 58:250–264
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP—the NCBI database of genetic variation. Nucleic Acids Res 29:308–311
Sunyaev S, Ramensky V, Bork P (2000) Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet 16:198–200
Sunyaev S, Lathe W, Bork P (2001) Integration of genome data and protein structures: prediction of protein folds, protein interactions and ‘molecular phenotypes’ of single nucleotide polymorphisms. Curr Opin Struct Biol 11:125–130
Xi T, Jones IM, Mohrenweiser HW (2004) Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics 83:970–979
Yuan H-Y, Chiou J-J, Tseng W-H, Liu C-H, Liu C-K, Lin Yi-Jung, Wang H-H, Yao A, Chen Y-T, Hsu C-N (2006) FASTSNP- an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res 34:635–641
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Namboori, P.K.K., Vineeth, K.V., Rohith, V. et al. The ApoE gene of Alzheimer's disease (AD). Funct Integr Genomics 11, 519–522 (2011). https://doi.org/10.1007/s10142-011-0238-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-011-0238-z