Abstract
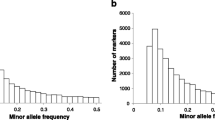
Genomic selection (GS) is an effective method to improve predictive accuracies of genetic values. However, high cost in genotyping will limit the application of this technology in some species. Therefore, it is necessary to find some methods to reduce the genotyping costs in genomic selection. Large yellow croaker is one of the most commercially important marine fish species in southeast China and Eastern Asia. In this study, genotyping-by-sequencing was used to construct the libraries for the NGS sequencing and find 29,748 SNPs in the genome. Two traits, eviscerated weight (EW) and the ratio between eviscerated weight and whole body weight (REW), were chosen to study. Two strategies to reduce the costs were proposed as follows: selecting extreme phenotypes (EP) for genotyping in reference population or pre-selecting SNPs to construct low-density marker panels in candidates. Three methods of pre-selection of SNPs, i.e., pre-selecting SNPs by absolute effects (SE), by single marker analysis (SMA), and by fixed intervals of sequence number (EL), were studied. The results showed that using EP was a feasible method to save the genotyping costs in reference population. Heritability did not seem to have obvious influences on the predictive abilities estimated by EP. Using SMA was the most feasible method to save the genotyping costs in candidates. In addition, the combination of EP and SMA in genomic selection also showed good results, especially for trait of REW. We also described how to apply the new methods in genomic selection and compared the genotyping costs before and after using the new methods. Our study may not only offer a reference for aquatic genomic breeding but also offer a reference for genomic prediction in other species including livestock and plants, etc.





Similar content being viewed by others
References
Ao J, Mu Y, Xiang L-X, Fan D, Feng M, Zhang S, Shi Q, Zhu L-Y, Li T, Ding Y (2015) Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet 11:e1005118
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Daetwyler HD, Villanueva B, Bijma P, Woolliams JA (2007) Inbreeding in genome-wide selection. J Anim Breed Genet 124:369–376
Duchemin S, Colombani C, Legarra A, Baloche G, Larroque H, Astruc J-M, Barillet F, Robert-Granié C, Manfredi E (2012) Genomic selection in the French Lacaune dairy sheep breed. J Dairy Sci 95:2723–2733
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Goddard ME, Hayes BJ (2007) Genomic selection. J Anim Breed Genet 124:323–330
Habier D, Fernando RL, Dekkers JC (2009) Genomic selection using low-density marker panels. Genetics 182:343–353
Henderson, C.R. (1975). Best linear unbiased estimation and prediction under a selection model. Biometrics 423–447
Legarra A, Robert-Granié C, Manfredi E, Elsen J-M (2008) Performance of genomic selection in mice. Genetics 180:611–618
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Liu X, Sui B, Wang Z, Cai M, Yao C, Chen Q (2011) Estimated reproductive success of brooders and heritability of growth traits for large yellow croaker (Larimichthys crocea) using microsatellites. Chin J Oceanol Limnol 29:990–995
Macciotta, N.P., Gaspa, G., Steri, R., Pieramati, C., Carnier, P. & Dimauro, C. (2009). Pre-selection of most significant SNPS for the estimation of genomic breeding values. BMC Proc 3(Suppl 1):S14
Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Meuwissen T (2009) Accuracy of breeding values of ‘unrelated’ individuals predicted by dense SNP genotyping. Genet Sel Evol 41:35
Meuwissen T, Goddard M (2010) Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185:623–631
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Meuwissen T, Solberg TR, Shepherd R, Woolliams JA (2009) A fast algorithm for BayesB type of prediction of genome-wide estimates of genetic value. Genet Sel Evol 41:2
Nielsen HM, Sonesson AK, Yazdi H, Meuwissen TH (2009) Comparison of accuracy of genome-wide and BLUP breeding value estimates in sib based aquaculture breeding schemes. Aquaculture 289:259–264
Ober U, Ayroles JF, Stone EA, Richards S, Zhu D, Gibbs RA, Stricker C, Gianola D, Schlather M, Mackay TF (2012) Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet 8:e1002685
Schulz-Streeck, T., Ogutu, J.O. & Piepho, H.-P. (2011). Pre-selection of markers for genomic selection. BMC Proc 5(Suppl 3):S12
Tsai H-Y, Hamilton A, Tinch AE, Guy DR, Gharbi K, Stear MJ, Matika O, Bishop SC, Houston RD (2015) Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics 16:969
Vanraden P (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Vanraden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel RD, Taylor JF, Schenkel FS (2009) Invited review: reliability of genomic predictions for North American Holstein bulls. J Dairy Sci 92:16–24
Wang Z, Wang Y, Lin L, Qiu S, Okamoto N (2001) Genetic polymorphisms in wild and cultured large yellow croaker Pseudosciaena crocea using AFLP fingerprinting. J Fish Sci China 9:198–202
Xiao S, Wang P, Zhang Y, Fang L, Liu Y, Li J-T, Wang Z-Y (2015) Gene map of large yellow croaker (Larimichthys crocea) provides insights into teleost genome evolution and conserved regions associated with growth. Sci Rep 5:18661
Yang X, Liu D, Liu F, Wu J, Zou J, Xiao X, Zhao F, Zhu B (2013) HTQC: a fast quality control toolkit for Illumina sequencing data. BMC Bioinformatics 14:33
Yang J, Jiang H, Yeh CT, Yu J, Jeddeloh JA, Nettleton D, Schnable PS (2015) Extreme-phenotype genome-wide association study (XP-GWAS): a method for identifying trait-associated variants by sequencing pools of individuals selected from a diversity panel. Plant J 84:587–596
Ye H, Liu Y, Liu X, Wang X, Wang Z (2014) Genetic mapping and QTL analysis of growth traits in the large yellow croaker Larimichthys crocea. Mar Biotechnol 16:729–738
Acknowledgments
Kun Ye, Qingkai Chen, Yang Liu, and other colleagues in the laboratory participated in fish sampling and the trait measurement. We thank two reviewers for providing many helpful suggestions.
Authors’ Contributions
ZW conceived and designed the study and revised manuscript; LD analyzed the data and drafted the manuscript; SX discovered SNPs and revised the manuscript; JC participated in the trait measurement and DNA extraction. LW participated in the genotyping costs estimation. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Statement
All fish were reared in a nucleus farm named “Jinling Aquaculture Science and Technology Co. Ltd.” in Ningde City, Fujian Province, P.R. China. The trail was carried out in the Key Laboratory of Healthy Mariculture for the East China Sea. This study was approved by the Animal Care and Use Committee of Fisheries College of Jimei University. All participants consented to publish the paper.
Availability of Data
Raw DNA sequencing reads were deposited in NCBI with the project accession of PRJNA309464 and SRA accession of SRR3114179.
Competing Interests
The authors declare that they have no competing interests.
Fundings
This work was supported by the National Natural Science Foundation of China (U1205122), Key projects of the Xiamen Southern Ocean Research Center (14GZY70NF34), the National ‘863’ Project of China (2012AA10A403), the Natural Science Foundation of Fujian Province (2016J05081) and the Foundation for Innovation Research Team of Jimei University (2010A02).
Rights and permissions
About this article
Cite this article
Dong, L., Xiao, S., Chen, J. et al. Genomic Selection Using Extreme Phenotypes and Pre-Selection of SNPs in Large Yellow Croaker (Larimichthys crocea). Mar Biotechnol 18, 575–583 (2016). https://doi.org/10.1007/s10126-016-9718-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-016-9718-4