Abstract
Receptor recognition is a crucial step in viral infection and is a critical factor for cell entry and tissue tropism. In several recent studies, angiotensin-converting enzyme 2 (ACE2) has been demonstrated to be the cellular receptor of SARS-CoV-2 as it was previously well known as the receptor of SARS-CoV. SARS-CoV-2 can bind with high affinity to human ACE2 and engages it as an entry receptor. It seems that the genetic, notably epigenetic variations of ACE2 are less known in different populations, indicating the need for its further investigation. These variations have the potential to affect its contribution to the pathogenicity of COVID-19. The contribution of epigenetics in the interindividual variability of ACE2 merits more attention because epigenetic processes can play important roles in ACE2 alterations in various tissues and different people and populations. Analyzing different DNA methylation patterns and microRNAs, contributing to the ACE2 modulation in the lungs will have a high priority. The epigenetic and genetic variations of ACE2 become even more important when considering that some people have mild clinical symptoms despite having COVID-19. The pathogenicity of SARS-CoV-2 infection is complex; therefore, a better understanding of the underlying pathobiology, especially binding the virus to its receptors, could help improve therapeutic and preventive approaches. This review aims to highlight the importance of evaluating both the epigenetic and genetic variations of ACE2 as a receptor for the deadly SARS-CoV-2.
Similar content being viewed by others
Introduction
The novel coronavirus disease 2019 (COVID-19) pandemic has become the most extraordinary global public health crisis since the pandemic influenza outbreak of 1918 [1]. Various aspects of this pandemic remain unknown. The outbreak of severe acute respiratory syndrome (SARS) in 2002–2003 uncovered the human body’s Achilles’ heel for SARS coronavirus (SARS-CoV), angiotensin-converting enzyme 2 (ACE2). Similarly, ACE2 is gaining importance for its identical role as the port of entry through which SARS-coronavirus 2 (SARS-CoV-2), also called the novel coronavirus 2019 (2019-nCoV), enters and hijacks cellular types of machinery for reproducing and debilitating the body's critical biological systems [2, 3]. While expressed in numerous tissues, the ACE2 is abundantly present in kidneys, liver, and cardiovascular system as well as the esophagus, stomach, intestines, bladder, testis, and brain; notably, the type II alveolar cells (AT2) in the lower respiratory tract possess the highest concentration of the ACE2 receptors [4,5,6,7]. This is consistent with the clinical outcomes of SARS-CoV-2 infection affecting and impairing these organs, sometimes to the extent of irreversible damage with eventual death [8, 9]. The new virus’s zoonotic origin highlights the hidden viruses in wild animals and their potential to occasionally spill over into humans [10].
ACE2 is a known negative regulator of the renin-angiotensin system (RAS), playing an essential part in blood pressure homeostasis and also a validated contributor in the pathogenesis of acute lung injury. ACE2 is the homolog of ACE whose function is to counterbalance ACE2 in the biochemical setting of RAS. While ACE cleaves angiotensin I (Ang I) into Ang II, a potent vasoconstrictor, ACE2 hydrolyzes Ang II into angiotensin- [1,2,3,4,5,6,7], a peptide with opposing effects [3, 11]. The ACE2 missing ACE2 knockout mice are severely impaired in cardiac functions, resulting in high blood pressure, irregular cardiac contractility, and cardiac hypertrophy or fibrosis [12, 13]. Besides, the ratio of ACE to ACE2 could be a potential prognostic indicator wherein their imbalance could result in hypertension [14, 15]. ACE2 has also been indicated to regulate other pathways such as the kinin-kallikrein system that plays an essential part in regulating inflammation and coagulation [16].
Intriguingly, the state of cell differentiation appears to affect the ACE2 surface expression, i.e., the more the cell’s differentiation, the higher the ACE2 expression [17]. Therefore, both the expression level and expression pattern of human ACE2 in various tissues may result in varied susceptibility, symptoms, and severity of SARS-CoV-2 infection. The evidence is further supported by the fact that far less or no pathogenicity was observed in SARS-CoV-2 infection in pediatric and teenage age groups, in contrast to the middle age and elderly with well-differentiated somatic cells [18].
Of note, the protective role of ACE2 expression against lung injury is downregulated by SARS-CoV-Spike (S). This executes ACE2 attenuation by modulating TNF-α-converting enzyme (TACE)-dependent shedding of the ACE2 ectodomain [19, 20]. There appears to be a likely scenario played out by the SARS-CoV-2-S for ACE2 downregulation in COVID-19. In addition, several lines of investigation suggested that the ACE2 receptor is the essential element for SARS-CoV infectivity and pathogenicity [21,22,23]. Also, they highlighted that the affinity between the viral receptor binding domain (RBD) and the host ACE2 during viral attachment is critical to dictate the range of hosts susceptible to SARS-CoV infection [23,24,25]. Importantly, research using various bat species provided evidence for varying genetic diversity of ACE2 gene as being the source of deciding which bats are likely to be the SARS-CoV host. The findings indicated that bat ACE2 receptor affinity to the RBD of SARS-CoV S protein and its efficiency could be modified by altering ACE2 crucial residues that were mostly consistent with structural modeling predictions [26].
Biophysical and structural evidence has shown that the binding capacity of SARS-CoV-2 spike binds ACE2 is much stronger than that of SARS-CoV spike [27, 28]. The SARS-CoV-2 spike glycoprotein binds to the peptidase domain (PD) of ACE2 with a Kd of ~ 15 nM, which is approximately 10- to 20-fold greater than the SARS-CoV-S protein does [27]. This high-affinity SARS-CoV-2 for human ACE2 is in agreement with the efficient transmission of SARS-CoV-2 in humans reported to date [29]. In other words, ACE2-binding affinity can be one of the most important determinants of SARS-CoV infectivity; thus, SARS-CoV-2 has evolved the capability to infect humans and to transmit among humans [23].
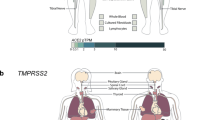
Variations in symptoms and incubation times and polymorphic severity could reflect both evolving mutant strains of SARS-CoV-2 and host genetic factors blended with environmental ones [30]. According to genetic evidence, up to 67% of the variability in circulating ACE2 concentrations may be explained by heritable factors [5]. Studies in different races and ethnicities showed that genetic variants of ACE2 were associated with hypertension, coronary heart disease, and some other cardiovascular events [31]. It was also demonstrated that higher nasal expression of transmembrane serine protease 2 (TMPRSS2) used by SARS-CoV-2 for S protein priming might be involved in the higher burden of COVID-19 in Black people [32]. Furthermore, epigenetics’ contribution in the intraindividual and interindividual variability of ACE2 may require more attention because epigenetic processes, in particular DNA methylation, can play an important role in ACE2 alterations in various tissues as well as different peoples. The importance of the epigenetic and genetic variations of ACE2 as the SARS-CoV-2 receptor becomes even more evident when considering that some people exhibit mild clinical symptoms despite having COVID-19. A better understanding of this new virus’s underlying pathobiology, notably its binding to the human receptors, can help efficiently improve therapeutic and preventive approaches. This review aimed to evaluate the epigenetic and genetic variations of ACE2 as a receptor for SARS-CoV-2 to provide insights into the molecular mechanisms that affect ACE2 levels and its potential clinical implications (Fig. 1).
An overview of the epigenetic and genetic variations of angiotensin-converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV-2. The epigenetic and genetic variations of ACE2 need to be considered together to better understand the various factors that contribute to regulating this crucial enzyme
Protein-protein interaction between ACE2 and SARS-CoV-2
ACE2 is a type I membrane protein of 805 amino acids composed of an N-terminal PD and a C-terminal Collectrin-like domain (CLD) that ends with a single transmembrane helix and a ~ 40-residue intracellular segment [11, 33]. In mammals, ACE2 is highly conserved in terms of the peptide sequence; that is, in all mammals, its protein exhibits a homology of 70% or more [34]. This protein is a zinc metallopeptidase. The active site of ACE2 contains the His-Glu-Met-Gly-His zinc-binding motif, which its residues are highly conserved [4]. The zinc-binding motif repeats twice in ACE and once in ACE2. ACE2 catalyzes enzymatic reactions by utilizing zinc ions, coordinated by conserved histidines within the motif [35].
Coronavirus entry into host cells is mediated by the envelope S glycoprotein, which is crucial for determining host tropism and transmission capacity. The spike protein of coronaviruses comprises two functional domains responsible for binding to the host cell receptor (S1 domain) and cell membrane fusion (S2 domain) [10, 29]. The S1 domain contributes to the stability of the prefusion state of the membrane-anchored S2 domain that contains the fusion machinery [29]. The receptor-binding domain (RBD) of beta coronaviruses is commonly located in the C-terminal domain of S1 [10]. The RBD of S1 directly binds to the PD of ACE2 [11]. When S1 binds to ACE2, another cleavage site on S2 is exposed and cleaved by host proteases, a critical process for viral infection [11]. It was shown that the new virus does not exploit other coronavirus receptors such as aminopeptidase N and dipeptidyl peptidase 4 [36]. Although the lengths of most of the proteins encoded by SARS-CoV-2 and related viruses were similar, a considerable difference was a longer spike protein encoded by SARS-CoV-2 (1273 amino acids) compared with bat-SL-CoVZC45 (1246 amino acids), bat-SL-CoVZXC21 (1245 amino acids), SARS-CoV (1255 amino acids), and MERS-CoV (1270 amino acids) [10].
The genome of SARS-CoV-2 shares about 80% identity to that of SARS-CoV [11]. Their spike proteins have 76.5% identity in amino acid sequences, and, importantly, these proteins have a high degree of homology [36]. The remarkable structural similarity and sequence conservation between the SARS-CoV-2 S and SARS-CoV S glycoproteins indicates the close relationship between these two viruses recognizing hACE2 to enter target cells [29]. Based on a comparative model for the ACE2-RBD interactions in SARS-CoV RBD and SARS-CoV-2 RBD, Wan and colleagues [23] suggested that SARS-CoV-2 has gained some capability to transmit from human to human.
Comparison of the amino acid identities between hACE2 and ACE2s of Chinese horseshoe bats, civet, swine, and mouse has indicated that mouse ACE2, which cannot be utilized by SARS-CoV-2, has higher similarity to hACE2 than those of Chinese horseshoe bats and swine, which can be used by SARS-CoV-2, showing that whole amino acid sequence identities cannot be a good criterion for predicting engaging capability by SARS-CoV-2 [37]. It was found that that the SARS-CoV-2 RBD sequence from 329 to 521 shares 93% identity with pangolin SARS-like CoV SRR10168377. Accordingly, it has been proposed that the pangolin′s virus has a higher probability of infecting humans [28]. Furthermore, aligning hACE2 with that of other species indicated that Lys31, Glu35, Asp/Glu38, and Lys353 are conserved and probably involved in forming salt bridges [38].
The overall interface between SARS-CoV-2 and ACE2 is similar to that between the SARS-CoV and ACE2, which seems to be mediated mainly through polar interactions. The RBD of the surface S glycoprotein of SARS-CoV-2 is recognized by the PD of ACE2 primarily through polar residues. Structural studies suggested that two S protein trimers can simultaneously bind to an ACE2 homodimer so that each PD accommodates one RBD [11].
Crucial amino acids in the binding of spike protein to ACE2
The modeling of the three-dimensional structure of the SARS-CoV-2 receptor-binding domain using the Swiss-Model program showed that, like other beta coronaviruses, the RBD was composed of a core and an external subdomain [10]. In particular, the external subdomain of the SARS-CoV-2 RBD was more similar to SARS-CoV. Lu et al. reported that key residues that account for the SARS-CoV RBD binding to the ACE2 receptor were variable in the SARS-CoV-2 RBD. These residues in the receptor-binding site of the SARS-CoV-2 RBD include Asn439, Gly485, Phe486, Gln493, and Asn501 [10]. Like these findings, Wan et al. [23] indicated that Asn501 and Phe486 of SARS-CoV-2 RBD enhance viral binding to human ACE2. Moreover, according to the structural models of Liu and colleagues [28], SARS-CoV-2 S protein may bind to ACE2 through Leu455, Phe486, Gln493, Asn501, and Tyr505. In the external subdomain of SARS-CoV, residues Leu472, Asn479, and Thr487 are the prominent interacting positions. Residues Asn479 and Thr487 in SARS-CoV correspond to Gln493 and Asn501 in SARS-CoV-2, respectively [28].
Detailed analysis of the interface between RBD and ACE2 by Yan et al. revealed that the PD of ACE2 mainly engages the α1 helix to recognize the RBD. The researchers divided the contact region between RBD and ACE2 into three clusters. At the amino terminus of α1, Gln498, Thr500, and Asn501 of the RBD form a network of hydrogen bonds with Tyr41, Gln42, Lys353, and Arg357 on ACE2. In the middle cluster, there was an interaction between Lys417 and Tyr453 of the RBD with Asp30 and His34 of ACE2, respectively. At the carboxyl terminus of α1, Gln474 of RBD is hydrogen-bonded to Gln24 of ACE2, while Phe486 of RBD interacts with Met82 of ACE2 through van der Waals forces [11].
Noteworthy, a closer look at the above studies shows that the residues Phe486 and Asn501 and as well as Gln493 have commonly been identified as key amino acids in the binding of the SARS-CoV-2 RBD to the hACE2 receptor and will therefore play a vital role in this binding.
Interestingly, certain significant changes occurring in the mouse and/or rat ACE2 in comparison to the human ACE2 may partially explain why mouse ACE2 does not mediate SARS-CoV-2 infection [38]. These data may also reveal what amino acids can be key in hACE2 to bind to SARS-CoV-2. The Asn31 (Lys31 in human), Ser82 (Met82 in human), and His353 (Lys353 in human) in mouse ACE2 may not form favorable interactions with SARS-CoV-2. Therefore, these changes may prove important in the binding affinity of S protein to ACE2. The changes into Asn31 and His353 in mouse ACE2 may affect the salt bridge formation as Lys31, and Lys353 do in hACE2. Also, the change into Ser82 with its hydrophilic property in mouse ACE2 may prevent the hydrophobic interaction of the original Met82. Consequently, these findings explain, in part, why mouse ACE2 does not mediate SARS-CoV-2 infection [38].
It is worth noting that finding the point mutations that affect the binding affinity between SARS-CoV-2 and hACE2 can be valuable. In this context, Korber et al. identified a crucial SARS-CoV-2 variant in the spike protein (D614G). They reported that more viruses circulating in the human population globally are the G614 form of the spike protein versus the D614 form that was initially identified from the first human cases in Wuhan, China. G614-bearing viruses have significantly higher infectious titers in vitro than their D614 form [39]. Wan et al. also reported that a single N501T mutation (corresponding to the S487T mutation in SARS-CoV) might significantly increase the binding affinity between SARS-CoV-2 RBD and human ACE2 [23].
Genetic variations of ACE2
In humans, located on chromosome Xp22, the ACE2 gene contains 18 exons spanning approximately 40 kb of genomic DNA coding for a protein of 805 amino acids [5, 11]. As one of the evolutionary sources for shaping genes and genomes, variations in the genomic structure of mammals have evolved during their complicated coexistence with viruses that alter interfaced proteins [40]. Therefore, since the interactions between the ACE2 receptor and the S-protein RBD domain of SARS-CoV-2 are critical for the virus’s cellular entry, ACE2 variations may decrease or increase its affinity to the S-protein RBD that may protect or render individuals more susceptible to the virus. Based on heritability analyses, the Leeds Family Study, Rice et al. [41] disclosed that genetic factors accounted for 67% of the phenotypic variation in circulating ACE2, while the amount was 24.5% for ACE. Genetic variations, therefore, in the gene encoding ACE2 can be a strong candidate for diversities in ACE2 function and its receptor role for SARS-CoV-2. Accordingly, Cao et al., by comparing the allele frequencies of the 15 variants of unique expression quantitative trait loci (QTL) among different populations, reported that the genotypes of ACE2 gene polymorphism might influence the expression levels of ACE2 in the populations. They also proposed that according to the differences in the allele frequencies of ACE2 coding variants among different populations, the genetic variations might influence ACE2 functions in populations. The researchers identified a singleton truncating variant of ACE2 (Gln300X) in the ChinaMAP. Furthermore, they introduced seven hotspot variants, including Lys26Arg, Ile468Val, Ala627Val, Asn638Ser, Ser692Pro, Asn720Asp, and Leu731Ile/Leu731Phe in different populations [42].
The role of regulatory elements in ACE2 expression
Given the factors and characteristics involved in regulating ACE2 expression, Kuan et al. reported a DNA sequence element, -516/-481, upstream of the ACE2 gene as a likely protein binding domain, which appears to be required for Ang II-activated transcription. They also claimed to discover a regulatory sequence, ATTTGGA, within the -516/-481 region of the ACE2 gene and suggested a role for the sequence to play in regulating ACE2 expression [43]. Also, using the human embryonic kidney cell line (HEK293), hepatocyte nuclear factor 1beta (HNF1β) was found to promote ACE2 transcription since it has functional HNF1β binding sites in its promoter region [44]. Pedersen et al. [45] further elaborate on the regulatory structure and functions of the ACE2 promoter in terms of its expression at the level of mRNA, protein, and enzymatic activity. They found three HNF1 binding motifs within the proximal ACE2 promoter region highly conserved among mammalian species. Both HNF1α and HNF1β indicated binding affinity to these three conserved elements acting cooperatively to mediate their promoting impact on ACE2 expression. Besides, neither HNF1α nor HNF1β displayed affinity to the distal promoter region, while a GATA binding motif was found in the distal promoter region. Since the distal promoter region could not act autonomously, its role was suggested to be in conjunction with the proximal promoter region to form a single functional domain to act on ACE2 gene.
The upstream promoter region of the human ACE2 gene has two distinct parts, -454/-1 and -1509/-928, conserved in placental mammals. The bipartite ACE2 promoter consists of distal and proximal regions separated with an Alu element in humans and short interspersed elements in the mouse. Yet, these two regions in ACE2 promoters of other placental mammals are adjoining [45]. Interestingly, the ACE2 gene, both in humans and mice, has at least two transcription start sites; hence, there exist two types of ACE2 mRNAs: (I) ACE2 mRNA with transcriptional initiation in the proximal promoter region yielding proximal promoter transcripts (PPT) and (II) ACE2 mRNA with transcriptional initiation in the proximal promoter region yielding distal promoter transcripts (DPT) both of which produce the same ACE2 polypeptide. Moreover, tissues expressing ACE2 display unbalanced concentrations of DPT and PPT. While both forms prevail in the kidney, pancreas, heart, and brain, PPT is more abundant than DPT in these organs, at least 2-fold.
On the contrary, only in the lung is the DPT expressed more than PPT [45]. In another study, Pedersen et al. identified other conserved cis-regulatory elements at positions -153/-144 and -101/-79 of the human ACE2 promoter that appears required for basal transcription. The forkhead box (FOX) transcription factor, FOXA2, was suggested to attach the -153/-144 motif, thereby inducing ACE2 transcription. Yet, the authors stressed the possibility of additional factors affecting both ACE2 and FOXA2 expression [46]. This is in line with the possible estrogen-induced ACE2 expression and elevated ACE2 enzymatic activity in female mice than males, even though it’s presumably cell and tissue-specific [47, 48]. Furthermore, the gene dosage effect is possibly a strong factor as the ACE2 gene is located on the X chromosome.
ACE2 polymorphisms related to SARS and SARS-CoV-2 infection
A pathogen receptor/ligand is a molecule through which it interacts with its cognate receptor/ligand on the host cell at an initial step of infection and thus is an indispensable factor for infection. Besides, the host cell’s viral receptor or co-receptor is a determinant factor in viral infection. On the other hand, different mice strain studies have suggested that genetic factors strongly affect the host’s response to pathogens and human studies illustrated by racial and familial differences in susceptibility to infection [49]. As a case in point, a mutant allele of genetic polymorphism in chemokine receptor 5 (CCR5), co-receptor of human immunodeficiency virus-1 (HIV-1), results in a dysfunctional protein which consequently renders host cells resistant to HIV-1 infection and thus changes its clinical course [50]. Moreover, it’s reported that the mouse hepatitis virus (MHV) receptor expressed in MHV-susceptible BALB/c mice exhibited approximately 10 to 30 times more binding affinity than that expressed on MHV-resistant mice cells [51]. Therefore, by analogy with the preceding findings, it is of foremost importance to consider the genetic variations and polymorphisms of ACE2 since they may potentially affect SARS-CoV-2 infection.
In accordance, two separate studies investigated and found several single nucleotide polymorphisms (SNPs) in the ACE2 gene concerning SARS infection, even though they ruled out the link between common genetic variants in the ACE2 gene and susceptibility to SARS. Chiu et al. identified 103 SNPs in the ACE2 gene, of which two were located within the coding regions, rs4646116 and rs4646179. The remaining ones were spotted within the introns of ACE2, and SNP validation confirmed sequence variations at five SNP loci that all were noncoding, rs2106809, rs2285666, rs4646142, rs714205, and rs2074192. Comparing the frequencies of these five polymorphisms among SARS patients and healthy controls of Chinese ethnicity, no significant difference was reported between the female and male controls in terms of allele distributions, between the SARS patients and controls, between SARS patients with poor outcomes and controls, together with pairwise gender-based comparisons [52]. A year later, Itoyama et al. published their investigation as to the previously known SNPs and their own newly found SNPs. They identified one novel non-synonymous substitution N638S plus 18 noncoding SNPs, of which 13 were new polymorphisms, and 3 out of 13 were located in 3′UTR. Taken together, no evidence was found concerning ACE2 polymorphism and SARS infection or its clinical course [50]. However, given the results which failed to establish a link between ACE2 polymorphisms and SARS infection, we should have thought of some points to consider: (1) likely insufficient sample size in these studies, (2) extending studies to explore variations in regulatory elements of ACE2, (3) paying attention to the stronger affinity between the spike protein of SARS-CoV-2 and ACE2 compared with the case in SARS and ACE2, and ultimately (4) in the context of COVID-19 and SARS-CoV-2, there is a co-receptor, TMPRSS2, which requires further genetic polymorphism studies coupled with ACE2.
Epigenetic regulation of ACE2
Epigenetics is the study of heritable changes in gene expression in the absence of DNA sequence changes [53]. Epigenetic mechanisms provide a molecular explanation to bridge the gap between the genome and environment, mediated by DNA methylation, histone modifications, noncoding RNA alterations such as long noncoding (lnc) RNAs and microRNAs (miRNAs, miRs), and chromatin folding proteins, which collectively, enable the cells to quickly respond to environmental stimuli [53, 54]. Epigenetics can create a new framework contributing to the search for etiological factors in complex diseases and could thus lead to targeted therapies. Epigenetic processes describe how a number of cells with the same genome can be differentiated into distinct cell types with different phenotypes. The increasing knowledge of epigenetics is altering our understanding of biology and medicine [55].
ACE2 and DNA methylation
DNA methylation is the addition of methyl groups to the cytosine residues in CpG dinucleotides. The majority of CpG dinucleotides in the mammalian genome are constitutively methylated. However, short interspersed DNA sequences diverge significantly from the average genomic pattern by being GC-rich, CpG-rich, and predominantly non-methylated, namely, the CpG islands [56, 57]. The risk factors of diseases can impose aberrant DNA methylation patterns, which change the methylation pattern in sequences normally hyper-, hypo, or un-methylated [58].
Of interest, CpG dinucleotide levels can be an essential factor for both the virus and the host antiviral defense. Different hosts or host tissues providing various antiviral activities may alter RNA modification signatures found in the viral RNA genome. The zinc-finger antiviral protein (ZAP) responsible for recognizing and binding CpG dinucleotides in viral genomes is able to recruit accessory proteins and degrade the targeted viral genomes. Indeed, increasing the CpG content of a viral genome, which evolved as a low CpG genome, could abort viral replication and subsequent virulence in response to ZAP activity. Interestingly, however, SARS-CoV-2 having the most extreme CpG deficiency among Betacoronavirus genomes may direct its host organism or tissue inhabitation in terms of ZAP activity to provide an environment with the highest selecting pressure against CpG. Thus, it seems critical to focus on its original hosts to figure out their specific selective milieu and viral defenses [59].
DNA methylation can be one of the important mechanisms of ACE2 regulation; however, it has not been fully addressed. In a study, Corley et al. analyzed four available genome-wide DNA methylation human datasets. They showed that DNA methylation among three CpGs of the ACE2 gene was lowest in lung epithelial cells than the other tissue cell types, suggesting that transcription and expression were highest in the lung/respiratory system. The findings of these researchers in airway epithelial cells indicated that DNA methylation near the transcription start site of the ACE2 gene was significantly associated with biological age [60]. Moreover, DNA methylation analysis at a CpG the ACE2 gene revealed a large degree of variability in both men and women. The effects of sex on ACE2 methylation were also shown by Fan et al. [61]. They analyzed the five CpG sites in the ACE2 promoter and revealed that CpG2 methylation was higher in males than in females, and conversely, CpG5 methylation was lower in males. In other words, sex may influence ACE2 methylation, which in turn may affect ACE2 levels. Also, according to the results by Cardenas et al., nasal ACE2 DNA methylation reflects differences by sex, race/ethnicity, and biological aging. They concluded that differences in nasal ACE2 DNA methylation could involve understanding COVID-19 severity and disparities reflecting upstream environmental and social effects [62]. In addition, Fan and colleagues reported that the methylation of some CpG sites in the ACE2 promoter might affect essential hypertension [61].
In a recent study, Sawalha et al. [63] provided evidence that ACE2 overexpression in the T cells of lupus patients is mediated through DNA methylation and may increase the risk of SARS-CoV-2 infection. The researchers introduced ACE2 as a methylation-sensitive gene located on the X chromosome. Based on whole-genome DNA methylation data, they found the hypomethylation of the ACE2 gene, which was mediated through several CpG sites located in the ACE2 promoter region proximal to the transcription start site, the 5’-UTR, and the 3’-UTR. Of interest, oxidative stress induced by viral infections can strengthen the DNA methylation defect in lupus, leading to further ACE2 hypomethylation and enhanced viremia. Therefore, hypomethylation and overexpression of ACE2 in T cells (and possibly other cells expressing ACE2) will facilitate T cell viral infections and viral distribution, which will contribute to susceptibility to COVID-19. Their findings propose that epigenetic dysregulation might elevate the risk and severity of SARS-CoV-2 infection in lupus patients [63].
ACE2 and histone modifications
The core histones in the nucleosomes are densely packed; however, histone-modifying enzymes can modify their tails. The counterbalancing works of various enzymes make epigenetic modifications reversible. Enzymes such as histone acetyltransferases (HATs) are balanced by histone deacetylases (HDACs), and histone methyltransferases (HMTs) are counteracted by histone demethylases (HDMs) in maintaining the epigenome [64, 65].
It was reported that silent information regulator T1 (SIRT1), a histone deacetylase, may be involved in the transcriptional regulation of ACE2 expression, and this role may be related to the protective role of SIRT1 against cellular stress [66]. In this context, the role of non-steroidal anti-inflammatory drugs as the inhibitors of SIRT1 deacetylase activity may also be considered [67]. Furthermore, drug-induced epigenetic changes should be noted in the Regulation of ACE2. For instance, based on the findings of Tikoo et al. [68] in high cholesterol diet rabbits, atorvastatin can upregulate ACE2 through promoter-specific histone modifications, which occur in a tissue-specific manner.
Recently, Pinto et al. [69] expressed that elevated ACE2 expression in the lungs of patients with comorbidities associated with severe COVID-19 may be due to genes related to histone modifications, such as HAT1, ADAC2, and lysine demethylase 5B (KDM5B). The authors reported that epigenetic marks in the ACE2 locus were compatible with those promoted by KDM5B. KDM5B can influence chromatin accessibility by removing active chromatin marks such as di- and tri-methylation from the lysine 4 of histone H3 (H3K4), contributing to gene transcription regulation and DNA repair. KDM5B inhibition in breast cancer cells was found to trigger an interferon response resulting in DNA and RNA viruses’ resistance to infection. This proposes that KDM5 demethylases may be potential targets for COVID-19 prevention [69].
It is worth noting that some natural compounds such as curcumin may induce epigenetic silencing of the ACE2 gene, showing a preventive potential towards COVID-19 infection [70]. It was also found that genistein treatment increased HAT1 expression, and as a result, this soy isoflavone may play a role in modulating ACE2 [71].
The microRNA regulation of ACE2
The role of microRNAs in the gene regulation of ACE2 is also worth considering. Fortunately, there has recently been growing documentation on the role of miRNAs in the Regulation of the ACE2 gene. miRNAs are a class of small, noncoding, and single-strand RNA molecules that work by post-transcriptionally inhibiting the expression of a large number of target genes in various organisms ranging from viruses to higher eukaryotes. These small RNAs work through complementary binding to the 3′-UTR and occasionally to the 5′-UTR or the coding regions of target mRNAs [72,73,74]. The miRNAs have been identified as prominent biomarkers and sometimes detectable several years before the disease appears [75]. The contribution of miRNAs in host-pathogen interactions during infection may also be important. Better clarification of these interactions might provide a more in-depth understanding of viral pathogenesis and the development of efficient antiviral therapies as previously indicated in several viruses, including herpesvirus, enterovirus, and hepatitis C. The importance of miRNAs becomes more apparent when noting that small miRNA-like small RNAs are produced during viral infections, as reported in H5N1 Influenza, Ebola virus, and HIV-1 [76].
Downregulation of ACE2 is associated with acute lung injury or acute respiratory distress syndrome (ARDS) induced by the avian influenza virus, SARS-CoV, and respiratory syncytial virus [73]. Interestingly, the downregulation of ACE2 protein expression may be mediated by miRNAs [73]. Based on Lu et al.’s findings, ACE2 mRNA and ACE2 protein levels were inhibited by miR-200c in both rat primary cardiomyocytes and, importantly, in human-induced pluripotent stem cell-derived cardiomyocytes [77]. Liu et al. found that viral RNA of avian influenza virus can induce the upregulation of miR-200c-3p, which in turn downregulates ACE2 protein expression. They also found markedly increased plasma concentrations of miR-200c-3p in severe pneumonia patients. Therefore, it seems that blocking the function of this miRNA may be a potential therapeutic target. In human lung epithelial cells, the downregulation of a lncRNA (lncGAS5) reduces ACE2 expression via enhancing miR-200c-3p, contributing to promoting the progression of ARDS [78]. The contribution of the ALT1 lncRNA in the regulation of ACE2 was reported by Li and colleagues [79]. Accordingly, the knocking-down of ALT1 and ACE2 as its direct target gene could efficiently diminish the expression of cyclin D1. It was also shown that KDM5B could indirectly influence ACE2/TMPRSS2 expression by reducing transcription of let-7e/mir-125a and mir-141/miR-200 miRNA families [80].
Lambert et al. [81] identified putative miRNA-binding sites in the 3′-UTR of the ACE2 transcript using in silico analysis and showed that miR-421 is a potential regulator of ACE2. They introduced miR-421 as a novel potential therapeutic target, which could be used to regulate ACE2 expression in disease. There also is a significant and inverse association between the levels of circulating miR-421 and the ACE2 expression in leukocytes of patients with chronic kidney disease [82]. ACE2 was validated as a target gene of miR-1246 in the study of Fang et al. This miRNA mediates lipopolysaccharide-induced pulmonary endothelial dysfunction and apoptosis in vitro and lung inflammation and permeability in vivo through targeting ACE2. Accordingly, miR-1246 may be a potential therapeutic target for ARDS [72].
The altered expression of ACE2 in chronic hypoxia can occur with the contribution of hypoxia-inducible factor 1α (HIF-1α) and let-7b. Under hypoxia, HIF-1α upregulates let-7b, which in turn downregulates ACE2 expression by directly targeting the coding sequence of ACE2. This process contributes to hypoxic pulmonary hypertension’s pathogenesis by the proliferation and migration of stimulating pulmonary artery smooth muscle cells. It is concluded that let-7b may serve as a potential clinical target for the treatment of hypoxic pulmonary hypertension [83]. It was also reported that let-7c-5p targeted ORF1ab gene in SARS-CoV-2, and it was contributed to H1N1 influenza A suppression by targeting its M1 protein [76].
Predicting miRNA–mRNA pairs within the host system at the infection with SARS-CoV showed that ACE2 is a target gene of miR-186, miR-93, and miR-200b. Impacts of exercise training on specific miRNAs targeting RAS genes showed that it could decrease miR-143 targeting ACE2 in the heart [84]. It was also reported that miR-125b is a negative regulator of ACE2 through which mediates high glucose-induced ROS production and apoptosis in HK-2 renal tubular epithelial cells [85]. In the mice, miR-429 was indicated to contain a binding site on 3′-UTR of ACE2 and may play a role in its Regulation [86]. Analyzing the expression of placental miRNAs in the different gestation stages indicated that ACE2 could be a target for several miRNAs such as miR-936, miR-4270, and miR-144-3p [87].
Recently, Demirci and Adan [76] examined the miRNA-mediated regulation of SAR-CoV-2 infection by predicting possible viral genes targeted by cellular miRNAs and host genes targeted by viral miRNAs. They identified several viral miRNAs that might target human genes and introduced likely virulent genes of SARS-CoV-2 targeted by host-cellular miRNAs. Accordingly, all viral genes, which are mainly responsible for viral biogenesis, entrance, replication, and infection, may be potentially targeted by human miRNAs [76].
It is worth noting that several miRNAs contributing to ACE2 modulation may be candidates to be served as a potential clinical target in pulmonary disorders, including miR-1246 [72], miR-200c-3p [73], miR-421 [81], and let-7b [83]. The role of these miRNAs is most prominent when we find out that the SARS-CoV-2 causes lung injury in thousands of patients with COVID-19 worldwide. Successful targeting of these miRNAs can be a significant challenge in clinical management. However, the use of antagomirs may be clinically considered. In this context, it was reported that an antagomir to miR-145 could represent an alternative approach to increase ADAM17 (A Disintegrin And Metalloproteases 17) activity. This miRNA downregulates ADAM17. ADAM17, as a metalloprotease significantly expressed in the lungs and heart, is contributed to promoting the shedding of ACE2, thereby may provide a strategy to interfere with SARS-CoV-2 entry into the cells [88] (Table 1).
Furthermore, in addition to the regulation of the ACE2 gene at the mRNA level, other regulation levels such as translational efficiency, protein modification, folding, or subcellular localization should be considered [89].
Conclusions
A better understanding of this new virus’s molecular pathobiology, notably its binding to the human receptors, can help efficiently improve therapeutic and preventive approaches. It is of importance to consider epigenetic marks existing in ACE2 locus in addition to genetic variations since they may affect SARS-CoV-2 infection. Hypomethylation and overexpression of ACE2 in cells expressing ACE2, such as T cells, may contribute to susceptibility to COVID-19. The genes related to histone modifications such as histone acetyltransferase 1 may play a role in elevated ACE2 expression in the lungs of patients with comorbidities associated with severe COVID-19. miRNAs contributing to ACE2 modulation may be candidates to be potential clinical targets in the pulmonary complications of COVID-19.
Data availability
Not applicable
References
Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB (2020) Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A Review. JAMA.
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell.
Sodhi CP, Wohlford-Lenane C, Yamaguchi Y, Prindle T, Fulton WB, Wang S et al (2018) Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am J Phys Lung Cell Mol Phys 314(1):L17–L31
Hamming I, Cooper ME, Haagmans BL, Hooper NM, Korstanje R, Osterhaus AD et al (2007) The emerging role of ACE2 in physiology and disease. J Pathol 212(1):1–11
Burrell LM, Harrap SB, Velkoska E, Patel SK (2013) The ACE2 gene: its potential as a functional candidate for cardiovascular disease. Clin Sci 124(2):65–76
Zou X, Chen K, Zou J, Han P, Hao J, Han Z (2020) Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med:1–8
Qi F, Qian S, Zhang S, Zhang Z (2020) Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506
Tuttolomondo D, Frizzelli A, Aiello M, Bertorelli G, Majori M, Chetta A (2020) Beyond the lung involvement in COVID-19 patients. A review. Minerva Med
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science.
Gurley SB, Allred A, Le TH, Griffiths R, Mao L, Philip N et al (2006) Altered blood pressure responses and normal cardiac phenotype in ACE2-null mice. J Clin Invest 116(8):2218–2225
Yamamoto K, Ohishi M, Katsuya T, Ito N, Ikushima M, Kaibe M et al (2006) Deletion of angiotensin-converting enzyme 2 accelerates pressure overload-induced cardiac dysfunction by increasing local angiotensin II. Hypertension. 47(4):718–726
Brosnihan KB, Neves LA, Chappell MC (2005) Does the angiotensin-converting enzyme (ACE)/ACE2 balance contribute to the fate of angiotensin peptides in programmed hypertension? Am Heart Assoc
Wakahara S, Konoshita T, Mizuno S, Motomura M, Aoyama C, Makino Y et al (2007) Synergistic expression of angiotensin-converting enzyme (ACE) and ACE2 in human renal tissue and confounding effects of hypertension on the ACE to ACE2 ratio. Endocrinology. 148(5):2453–2457
Scialo F, Daniele A, Amato F, Pastore L, Matera MG, Cazzola M et al (2020) ACE2: the major cell entry receptor for SARS-CoV-2. Lung.:1–11
Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J et al (2005) ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 79(23):14614–14621
Sun J, He W-T, Wang L, Lai A, Ji X, Zhai X et al (2020) COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med
Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B et al (2005) Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 436(7047):112–116
Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T et al (2008) Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc Natl Acad Sci 105(22):7809–7814
Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S et al (2005) Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 24(8):1634–1643
Mahrooz A, Muscogiuri G, Buzzetti R, Maddaloni E (2021) The complex combination of COVID-19 and diabetes: pleiotropic changes in glucose metabolism. Endocrine. 2021/04/22
Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94(7)
McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L et al (2007) Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81(2):813–821
Yang X-L, Hu B, Wang B, Wang M-N, Zhang Q, Zhang W et al (2016) Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol 90(6):3253–3256
Hou Y, Peng C, Yu M, Li Y, Han Z, Li F et al (2010) Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch Virol 155(10):1563–1569
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O et al (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 367(6483):1260–1263
Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H et al (2020) Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol
Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell.
Mirmohammadi S, Kianmehr A, Arefi M, Mahrooz A (2020) Biochemical parameters and pathogenesis of SARS-CoV-2 infection in vital organs: COVID-19 outbreak in Iran. New Microbes New Infect 100792
Fan Z, Wu G, Yue M, Ye J, Chen Y, Xu B et al (2019) Hypertension and hypertensive left ventricular hypertrophy are associated with ACE2 genetic polymorphism. Life Sci 225:39–45
Bunyavanich S, Grant C, Vicencio A (2020) Racial/ethnic variation in nasal gene expression of transmembrane serine protease 2 (TMPRSS2). Jama. 324(15):1567–1568
Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T et al (2004) ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem 279(17):17996–18007
Jia H (2016) Pulmonary angiotensin-converting enzyme 2 (ACE2) and inflammatory lung disease. Shock. 46(3):239–248
Kuba K, Imai Y, Penninger JM (2013) Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ J 77(2):301–308
Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS (2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med:1–5
Qiu Y, Zhao Y-B, Wang Q, Li J-Y, Zhou Z-J, Liao C-H et al (2020) Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect
Li R, Qiao S, Zhang G (2020) Analysis of angiotensin-converting enzyme 2 (ACE2) from different species sheds some light on cross-species receptor usage of a novel coronavirus 2019-nCoV. J Infect
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W et al (2020) Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182(4):812–27.e19
Daugherty MD, Malik HS (2012) Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700
Rice GI, Jones AL, Grant PJ, Carter AM, Turner AJ, Hooper NM (2006) Circulating activities of angiotensin-converting enzyme, its homolog, angiotensin-converting enzyme 2, and neprilysin in a family study. Hypertension. 48(5):914–920
Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X et al (2020) Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov 6(1):1–4
Kuan T-C, Yang T-H, Wen C-H, Chen M-Y, Lee I-L, Lin C-S (2011) Identifying the regulatory element for human angiotensin-converting enzyme 2 (ACE2) expression in human cardiofibroblasts. Peptides. 32(9):1832–1839
Senkel S, Lucas B, Klein-Hitpass L, Ryffel GU (2005) Identification of target genes of the transcription factor HNF1β and HNF1α in a human embryonic kidney cell line. Biochim Biophys Acta (BBA)-Gene Struct Expr 1731(3):179–190
Pedersen KB, Chhabra KH, Nguyen VK, Xia H, Lazartigues E (2013) The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim Biophys Acta (BBA)-Gene Regul Mech 1829(11):1225–1235
Pedersen KB, Chodavarapu H, Lazartigues E (2017) Forkhead box transcription factors of the FOXA class are required for basal transcription of angiotensin-converting enzyme 2. J Endocr Soc 1(4):370–384
Wang Y, Shoemaker R, Thatcher SE, Batifoulier-Yiannikouris F, English VL, Cassis LA (2015) Administration of 17β-estradiol to ovariectomized obese female mice reverses obesity-hypertension through an ACE2-dependent mechanism. Am J Physiol-Endocrinol Metab 308(12):E1066–E1E75
Pedersen KB, Chodavarapu H, Porretta C, Robinson LK, Lazartigues E (2015) Dynamics of ADAM17-mediated shedding of ACE2 applied to pancreatic islets of male db/db mice. Endocrinology. 156(12):4411–4425
Malo D, Skamene E (1994) Genetic control of host resistance to infection. Trends Genet 10(10):365–371
Itoyama S, Keicho N, Hijikata M, Quy T, Phi NC, Long HT et al (2005) Identification of an alternative 5′-untranslated exon and new polymorphisms of angiotensin-converting enzyme 2 gene: Lack of association with SARS in the Vietnamese population. Am J Med Genet A 136(1):52–57
Ohtsuka N, Taguchi F (1997) Mouse susceptibility to mouse hepatitis virus infection is linked to viral receptor genotype. J Virol 71(11):8860–8863
Chiu RW, Tang NL, Hui DS, Chung GT, Chim SS, Chan KA et al (2004) ACE2 gene polymorphisms do not affect outcome of severe acute respiratory syndrome. Clin Chem 50(9):1683–1686
Cavalli G, Heard E (2019) Advances in epigenetics link genetics to the environment and disease. Nature. 571(7766):489–499
Ding N, Maiuri AR, O’Hagan HM (2019) The emerging role of epigenetic modifiers in repair of DNA damage associated with chronic inflammatory diseases. Mutat Res/Rev Mutat Res 780:69–81
Mahrooz A, Mackness M, Bagheri A, Ghaffari-Cherati M, Masoumi P (2019) The epigenetic regulation of paraoxonase 1 (PON1) as an important enzyme in HDL function: The missing link between environmental and genetic regulation. Clin Biochem
Meehan RR, Thomson JP, Lentini A, Nestor CE, Pennings S (2018) DNA methylation as a genomic marker of exposure to chemical and environmental agents. Curr Opin Chem Biol 45:48–56
Mahna D, Puri S, Sharma S (2018) DNA methylation signatures: biomarkers of drug and alcohol abuse. Mutat Res/Rev Mutat Res 777:19–28
Lund G, Zaina S (2009) Atherosclerosis risk factors can impose aberrant DNA methylation patterns: a tale of traffic and homocysteine. Curr Opin Lipidol 20(5):448–449
Xia X (2020) Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol Biol Evol
Corley MJ, Ndhlovu LC (2020) DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences
Fan R, Mao SQ, Gu TL, Zhong FD, Gong ML, Hao LM et al (2017) Preliminary analysis of the association between methylation of the ACE2 promoter and essential hypertension. Mol Med Rep 15(6):3905–3911
Cardenas A, Rifas-Shiman SL, Sordillo JE, DeMeo DL, Baccarelli AA, Hivert M-F et al (2020) DNA methylation architecture of the ACE2 gene in Nasal Cells. Medrxiv.
Sawalha AH, Zhao M, Coit P, Lu Q (2020) Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin Immunol 108410
Ordovás JM, Smith CE (2010) Epigenetics and cardiovascular disease. Nat Rev Cardiol 7(9):510
Vaissière T, Sawan C, Herceg Z (2008) Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res/Rev Mutat Res 659(1-2):40–48
Clarke NE, Belyaev ND, Lambert DW, Turner AJ (2014) Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin Sci 126(7):507–516
Dell’Omo G, Crescenti D, Vantaggiato C, Parravicini C, Borroni AP, Rizzi N et al (2019) Inhibition of SIRT1 deacetylase and p53 activation uncouples the anti-inflammatory and chemopreventive actions of NSAIDs. Br J Cancer 120(5):537
Tikoo K, Patel G, Kumar S, Karpe PA, Sanghavi M, Malek V et al (2015) Tissue specific up regulation of ACE2 in rabbit model of atherosclerosis by atorvastatin: Role of epigenetic histone modifications. Biochem Pharmacol 93(3):343–351
Pinto BG, Oliveira AE, Singh Y, Jimenez L, Goncalves AN, Ogava RL et al (2020) ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. medRxiv
Pruimboom L (2020) Methylation pathways and SARS-CoV-2 lung infiltration and cell membrane-virus fusion are both subject to epigenetics. Front Cell Infect Microbiol 10:290
Phillip CJ, Giardina CK, Bilir B, Cutler DJ, Lai Y-H, Kucuk O et al (2012) Genistein cooperates with the histone deacetylase inhibitor vorinostat to induce cell death in prostate cancer cells. BMC Cancer 12(1):145
Fang Y, Gao F, Hao J, Liu Z (2017) microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. Am J Transl Res 9(3):1287
Liu Q, Du J, Yu X, Xu J, Huang F, Li X et al (2017) miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov 3(1):1–17
Borghini A, Andreassi MG (2018) Genetic polymorphisms offer insight into the causal role of microRNA in coronary artery disease. Atherosclerosis. 269:63–70
Guay C, Regazzi R (2013) Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 9(9):513
Demirci MDS, Adan A (2020) Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. bioRxiv
Lu D, Chatterjee S, Xiao K, Riedel I, Wang Y, Foo R et al (2020) MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J Mol Cell Cardiol 148:46–49
Li H, Zi P, Shi H, Gao M, Sun R (2018) Role of signaling pathway of long non-coding RNA growth arrest-specific transcript 5/microRNA-200c-3p/angiotensin converting enzyme 2 in the apoptosis of human lung epithelial cell A549 in acute respiratory distress syndrome. Zhonghua Yi Xue Za Zhi 98(41):3354–3359
Li W, Wang R (2017) Ma J-y, Wang M, Cui J, Wu W-b, et al. A human long non-coding RNA ALT1 controls the cell cycle of vascular endothelial cells via ACE2 and cyclin D1 pathway. Cell Physiol Biochem 43(3):1152–1167
Nersisyan S, Shkurnikov M, Turchinovich A, Knyazev E, Tonevitsky A (2020) Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS One 15(7):e0235987
Lambert DW, Lambert LA, Clarke NE, Hooper NM, Porter KE, Turner AJ (2014) Angiotensin-converting enzyme 2 is subject to post-transcriptional regulation by miR-421. Clin Sci 127(4):243–249
Trojanowicz B, Imdahl T, Ulrich C, Fiedler R, Girndt M (2019) Circulating miR-421 targeting leucocytic angiotensin converting enzyme 2 is elevated in patients with chronic kidney disease. Nephron. 141(1):61–74
Zhang R, Su H, Ma X, Xu X, Liang L, Ma G et al (2019) MiRNA let-7b promotes the development of hypoxic pulmonary hypertension by targeting ACE2. Am J Phys Lung Cell Mol Phys 316(3):L547–LL57
Fernandes T, Hashimoto NY, Magalhães FC, Fernandes FB, Casarini DE, Carmona AK et al (2011) Aerobic exercise training–induced left ventricular hypertrophy involves regulatory MicroRNAs, decreased angiotensin-converting enzyme-angiotensin II, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1-7). Hypertension. 58(2):182–189
Huang Y, Zhang Y, Liu C, Huang J, Ding G (2016) microRNA-125b contributes to high glucose-induced reactive oxygen species generation and apoptosis in HK-2 renal tubular epithelial cells by targeting angiotensin-converting enzyme 2. Eur Rev Med Pharmacol Sci 20(19):4055
Goyal R, Van-Wickle J, Goyal D, Longo LD (2015) Antenatal maternal low protein diet: ACE-2 in the mouse lung and sexually dimorphic programming of hypertension. BMC Physiol 15(1):2
Wang Y, Lumbers ER, Arthurs AL (2018) Corbisier de Meaultsart C, Mathe A, Avery-Kiejda KA, et al. Regulation of the human placental (pro) renin receptor-prorenin-angiotensin system by microRNAs. MHR: Basic Sci Reprod Med 24(9):453–464
Rizzo P, Dalla Sega FV, Fortini F, Marracino L, Rapezzi C, Ferrari R (2020) COVID-19 in the heart and the lungs: could we “Notch” the inflammatory storm? Basic Res Cardiol 115(3)
Chen Y, Shan K, Qian W (2020) Asians do not exhibit elevated expression or unique genetic polymorphisms for ACE2, the cell-entry receptor of SARS-CoV-2. Preprints
Acknowledgements
The authors thank Mazandaran University of Medical Sciences as well as Golestan University of Medical Sciences. We wish to express our deepest gratitude to Prof. Giuseppe Poli (University of Turin, Italy) for his helpful comments and contributions.
Author information
Authors and Affiliations
Contributions
AM and AK were major contributors to writing the manuscript. AM, AK, IF, and OK checked and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kianmehr, A., Faraoni, I., Kucuk, O. et al. Epigenetic alterations and genetic variations of angiotensin-converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV-2: potential clinical implications. Eur J Clin Microbiol Infect Dis 40, 1587–1598 (2021). https://doi.org/10.1007/s10096-021-04264-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04264-9