Abstract
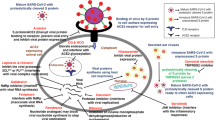
COVID-19 immunity in infected individuals may not be persistent. The specific response wanes in patients who have recovered from this infection. Nevertheless, it has not been fully understood whether true re-infection occurs or the viral reactivation. In this study, we investigated three COVID-19 patients who represented the symptoms after recovery. Chest CT scan was applied to assess the patients along with the viral samples from oropharyngeal/nasopharyngeal which were subjected to RT-PCR. The viral genome sequencing was applied where possible to distinguish possible re-infection or latent reactivation. Moreover, COVID-19-specific antibodies available data were evaluated in each incidence. The second episode of SARS-CoV-2 infection was different among the investigated subjects who experienced an interval between positive PCR tests ranged between 63 and 156 days. The disease presentation was less or more severe in the second infection. All cases were found IgG positive in the re-infection phase. The sequencing of SARS-CoV-2 sample obtained from two cases revealed a D614G mutation of S gene from the second isolated sample strengthens the case for the re-infection. The possibility of re-infection and reactivation could have significant effect on clinical implications and also vaccination. Our data supports clear warning of SARS-CoV-2 continuous circulation potency among the populations in spite of herd immunity either with natural infection or vaccination. This issue is critical in term of the patients, clinical investigate, and viral transmission.
Similar content being viewed by others
Introduction
COVID-19 pandemic has affected over 90 million infected people with more than two million deaths worldwide [1]. The high contagion of SARS-CoV-2 by droplets and other contact routes has resulted in outbreaks [2].
According to the studies which have been conducted on people who once caught COVID-19, a healthy immune response developed with both immunity arms, antibodies and T cells [3,4,5,6].
After the clearance of SARS-CoV-2 virus, the immune response is supposed to block a second infection due to generated memories against the virus. Considering this fact, people who recover from the infection should be protected from another possible infection for at least some amount of time; however, the length of this time has not been identified, yet [7, 8]. Some other viruses, the first infection could result in lifelong immunity, however, short-lived immunity is the outcome of seasonal coronaviruses [9, 10].
The exact mechanism of provided protection against SARS-CoV-2 has not been known, nor have the required levels of humoral or cellular responses been fully understood. Some probable factors associated with the re-infection are age, the health level of immunity system, correlated antibody, and possible virus mutations. [11].
Some studies have also shown that COVID-19 with mild presentation induces weaker immune responses which might provide short protection rather than severe form of the infection [12, 13]. Most infected SARS-CoV-2 individuals reach an antibody response during day 10 to day 21 post-infection. Mild cases display this response after 4 weeks, and in a minority of cases, IgM and IgG are not detectable [14,15,16]. Neutralizing antibodies are generated after infection, and their titers start to decline 2 months after the acute phase of the infection according to some researches. On the other hand, some studies detected antibodies for 6 months after the infection [17,18,19]. The second infection could be as the result of antibody faint or the viral sequence mutation especially in spike gene.
SARS-CoV-2 reactivation cases data are limited. COVID-19 therapy with curative and eradicative effect is not currently available. Reactivation seems to be more probable in those who suffers from an underlying condition or immune suppression. Subjects who are susceptible to reactivation are those with compromised immune systems such as dialysis patients or patients undergoing immune suppressive therapy [20, 21].
Therefore, fundamental question among the present pandemic is how long the immune responses against SARS-CoV-2 protect the host from re-infection or reactivation [9]. Moreover, a key question for COVID-19 is whether true re-infection occurs or not. It is really important to be sure about the negative cases in term of the test accuracy which brings the concern of SARS-CoV-2 reactivation in cases who got the infection and recovered. Moreover, the re-infection of the disease in the number of reported cases raises the consideration of vaccine efficacy against COVID-19 [22, 23].
Hereby, we report a case series of patients with COVID-19 who experienced reactivation or re-infection after negative PCR tests.
Materials and methods
In this study, COVID-19 suspected re-infected cases who were referred to COVID-19 National Reference laboratory at Pasteur Institute of Iran were investigated. COVID-19 was assessed upon admission based on the COVID-19 diagnosis and treatment flowchart in Iran (3rd edition) [24]. The oropharyngeal/nasopharyngeal swab specimens, in viral transport media (VTM), were assessed in the laboratory. Viral RNA was extracted using a QIAcube HT system with a QIAamp 96 Virus QIAcube HT Kit, according to the manufacturer’s instructions. Real-Time Reverse Transcription PCR (Real Time RT-PCR) assay was performed using 2019-nCoV Nucleic Acid Diagnostic kit (Sansure biotech, Changsha, China), according to the manufacturer’s protocol. Serology tests were also employed to assess humoral responses SARS-CoV-2 IgM Capture kit (cat no: PT-CoV-2-IgM Cap-96 [sensitivity 70%, specificity: 75%] Pishtazteb, Iran) and SARS-CoV-2 IgG kit (cat no: PT-CoV-19 IgG-96, [sensitivity 78%, specificity: 91%] Pishtazteb, Iran) were applied according to the provided protocol. The viral sample from each patient was finally subjected to sequencing to assess probable mutation. Partial N (466bp) and S (899bp) fragments were amplified using RT-PCR one step RT-PCR Kit (biotechrabbit, Germany) according to Table 1. All PCR products were analyzed by gel electrophoresis and were mostly confirmed by bidirectional Sanger sequencing. Due to the low quality of samples, we could only sequence it partially.
Patients’ confirmed recovery was described as two RT-PCT tests with negative result with 24-h interval. These mentioned methods were applied the same for both admission steps.
Case presentations
All the procedures were in accordance with the ethical standards of the responsible committee on human experimentation of Pasteur Institute of Iran under IR.PII.REC.1399.009 ethical code and also with the Helsinki Declaration.
Three COVID-19 confirmed cases by CT scan or RT-PCR in Pasteur Institute of Iran in March and April 2020, who presentedCOVID-19 symptoms some months after negative RT-PCR were studied (Tab.2). The patients’ symptoms resolution was achieved in all cases before the second symptoms initiation. None of the investigated subjects had underlying disease. Most importantly, case#1 had exposed to a confirmed infected family member and case#3 had contact to a confirmed colleague at workplace before they caught the disease in both episodes of the incidence. Only case #2 was not exposed to any confirmed patient. Furthermore, they did not catch the infection from the same source in each incidence and all cases fully recovered after the second episode.
The first case, a 32-year old woman, had an interval of 63 days between the two infections. The COVID-19 infection was confirmed by bilateral and peripheral ground-glass opacities in chest CT. The antibody titration was achieved positive by the rapid test (sensitivity 72%, specificity: 76%) for IgM (At the time of second infection, IgG titration was assessed as 4.89 AU/ml which after two months turned to a significant raise (over ELISA reader standard range). The disease presentation was similar in both incidences, however with more severe symptoms in the second time. The viral sequencing revealed a D614G mutation of S gene from the second isolated sample (Fig. 1).
The second case, a 54-year-old man whose infection was confirmed by RT-PCR. He recovered from the disease after 22 days with negative RT-PCR. After an interval of 156 days, the symptoms of the second infection appeared with less severity and gastrointestinal presentation. IgM and IgG were detected in the first incidence, and he was being followed up to the second virus presentation. In the whole duration between two incidences, IgG test was positive. Antibody titration at the time of second infection showed that IgG level was 5.25 IU/ml which increased to 27.5 IU/ml after about 2 weeks. The viral sequencing was obtained for N protein which showed no difference of the virus clade in both incidences (Fig. 2). Both sequences had L139L non-synonymous mutation, novel mutation in nucleotide position 28688. S portion sequencing was not achieved due to the low viral load.
A 42-year man was the third subject whose infection was confirmed by RT-PCR and bilateral and peripheral ground-glass opacities chest CT. He was fully recovered after the first infection and his RT-PCR was negative after 22 days. However, the antibody titration was not applied. He showed the second COVID-19 infection with 111 days of interval. The IgG titration was 17.5 IU/ml which decreased to 6.5 IU/ml after almost 2 weeks. His clinical symptoms were similar with more severe diarrhea in the second presentation step. In this case, the cycle threshold (Ct) was high, so the viral sequencing was not achieved. Moreover, the viral sequencing also revealed a D614G mutation of S gene from the second obtained sample (Fig. 1).
The serology tests of the cases were also investigated, and all three cases were found IgG positive at the re-infection incidence.
Discussion
The level of protective immunity mounted by SARS-CoV-2 infection is not currently known. Therefore, the re-infection probability has been found notably considerable. In this study, we reported 3 cases with suspicious for COVID-19 re-infection. The serology tests revealed that specific SARS-CoV-2 IgG presence is not enough to protect the second viral infection according to the findings. The disease presentation could be less or more severe in the re-infected patients. Moreover, the symptoms might be similar or different in each incident.
Recently, there have been few re-infected COVID-19 case reports. The first case was reported from Hong Kong, a 33-year-old man, who caught COVID-19 firstly in March with mild symptom presentation. After 4 months and a half, he was found PCR positive with high viral load but asymptomatic. Antibody seroconversion was recorded on the second infection. This might be according to the short-lived immunity which faints after a while [25].
The other reported case, a 25-year-old man from Nevada, the USA was confirmed to be infected with the SARS-CoV-2 twice. The interval between two infections was found 48 days. Surprisingly, he developed more serious disease in the second time including myalgia, cough, and shortness of breath and he required oxygen support. The data shows that both patients’ samples had genetic discordance which highlights the re-infection possibility [26].
Another case report from Brazil was a nurse who caught COVID-19 infection first in May 2020 with no specific underlying disease but only sporadically headache. Following the recovery and 38 days with no symptoms, late in June 2020, she faced severe headache, malaise, myalgia, fatigue, and fever feeling which developed with diarrhea and coughing. Her SARS-CoV-2 RT-PCR test was positive on 5th day of the symptoms. Nevertheless, the SARS-CoV-2 antibody detection rapid test (IgG/IgM) was negative. IgG was detected on 19th and 33rd days after the new onset of symptoms [27].
Another case report was a 20-year-old woman from Israel, diagnosed with Covid-19 in April 2020. She presented mild symptoms including fever and cough, with no respiratory distress and was confirmed by a positive nasopharyngeal PCR test. She cleared the virus in May. After 3 months, she became positive in August although she had no symptoms. Viral serology in August revealed positive SARS-Covid-2 IgG antibodies [28].
In the case report from Ecuador, the genome sequencing was found different in two incidents. The subject firstly presented mild infection in May, whereas the re-infection led to a moderate presentation [28, 29]. SARS-CoV-2 re-infection with worse presentation which results in oxygen support requiring and hospitalization raises concerns re-infection [30].
The USA and Ecuadorboth reportedre-infection cases who experienced increased symptom severity during second infection. The infected cases from Belgium, Hong Kong, and the Netherlands did not present more severe symptoms. There have been three possible explanations for the patient’s more serious symptoms including a higher viral load in the re-infection phase than the first infection, a more virulent version of the virus as the cause of the second incident and the developed antibodies which could result in the subsequent worse infection [30, 31].
In the study from Hong Kong, after two genome sequencing on respiratory specimens during two episodes of a COVID-19 infected patients, the results showed that the firstly viral genome and the second one were belonged to different virus clades with a stop codon at position 64 of orf 8 which led to a truncation of 58 amino acids. They concluded that serological and genomic analyses were strong clues for the re-infection instead of persistent viral shedding. The patient from Hong Kong did not develop Covid-19 antibodies in first disease, whereas the second presentation led to antibodies detection. The viral genomes of these studies from Hong Kong, Nevada, and Ecuador were sequenced to determine that the two infections as separate events which resulted differently [28, 29, 32, 33]. Coppolla et al. reported a PCR-confirmed COVID-19 patient who experienced viral reactivation with milder symptoms after 43-day interval and negative PCR. This case had dyslipidemia and chronic ischemic heart disease as underlying diseases [20].
The other SARS-CoV-2 reactivation case was a 55-year-old female suffering from Philadelphia chromosome-positive, CD20-positive B-ALL, asthma, coronary artery disease, and diabetes. After two negative PCR tests and discharge, she re-presented the infection with more severe symptoms. Considering the short time frame, reactivation seems more rational than re-infection. This highlights the risks of recently recovered COVID-19 patients who are on the treatment of immunosuppressive therapy and raises critical questions about the reactivation of SARS-CoV-2 [34]. Based on the observation, they suggested that recovered patients must be considered as the carriers of the virus and therefore an additional round of follow up and isolation are needed [20].
There was a reported case from Italy, who recovered from COVID-19 with positive serology. This 69-year-old woman was suffering from type 2 diabetes mellitus and urinary tract neoplasm. She was followed up for 1 month with six negative PCR tests. Nevertheless, she presented a second IgM seroconversion along with a positive RT-PCR after exposure to the virus [35].
More recently, new SARS-CoV-2 variant in Brazil has reported after the two mutated strains of the virus which were discovered in the UK and South Africa. The newly reported variant of the virus belongs to the B.1.1.248 strain which contains 12 mutations in the spike protein. Although viruses are naturally supposed to mutate, the SARS-CoV-2 variants are shown to be more transmissible in comparison with the original which started the pandemic. Therefore, this could result in higher numbers of serious infections and probable additional deaths [36].
Among the investigated cases in our study, sequencing also suggests that two cases, No#1 & No#3, infected in two separated steps according to the viral mutation (Fig. 1). The amino acid change in the spike protein of the virus, D614G, has emerged early during the pandemic as the viruses harboring this mutation are now dominant in many places worldwide [37]. There have been some evidences that the rapid spread of G614 was due to its more infectious potency than D614 and a higher levels of viral RNA has been assessed in the clinical samples from G614 infections[38, 39].
The recent study by usage of pseudovirus showed that 7% of convalescent sera obtained from recovered COVID-19 subjects contained reduced serum neutralizing activity against 614G in comparison with 614D [38]. According to the time of the first infection, the predominant circulating virus in Iran was D614 which initiated from China. After few months the G614 type was also found in Iran from the European countries. Therefore, we can assume that the second virus was different in the second infection occurrence and re-infection was probable occurrence. Moreover, the symptoms were more serious in the second incidence.
The other case, patient No#2, experienced 157 days of interval between the two incidences and was confirmed negative by RT-PCR. This patient was being followed up in every scene of the disease until he fully recovered. Interestingly, the IgG test was detected monthly in the period between to incidences. Although IgG level raised in the second infection and the symptoms appeared again, the virus type was the same in both infections suggesting that re-activation of the infection occurred (Table 2). As Fig. 2 shows, the N sequencing shows a non-sense mutation T:C based on the similarity between the symptoms and low severity in the second infection, reactivation seems more logical in this case. With the experience of two infection episodes by two SARS-CoV-2 positive specimens separated by a period of 157 days and also the resolution of symptoms with negative RT-PCR, reactivation incidence was more probable.
There has recently been an increasing number of COVID-19 patients with the second episode of the infection in Iran. It seems that the patients who experience the second infection have high Ct and low viral load. The other important issue is that all the investigated cases in this study were IgG positive at the second infection time with no specific underlying disease. Although the applying serological platforms are different worldwide and the comparison is not rational, the protection potency of IgG is the fundamental matter. The patients from the USA and Ecuador who had more serious symptoms in re-infection phase strongly highlight this issue. In other cases that experience mild presentation in the second episode, it is not fully understood that present antibodies play a role or not.
Conclusion
We presented three cases of possible SARS-CoV-2 re-infection or re-activation of latent virus, based on clinical and laboratory evidence.
The reported cases from different countries has raised the concern that re-infection or reactivation of SARS-CoV-2 affect the efficacy of emerging vaccines. It had been generally thought that once the infection occurs, immune response could prevent a second infection in the same person. Nevertheless, the possibility of re-infection which seems is growing worldwide has significant effect on clinical implications and also vaccination. The serology tests revealed that specific SARS-CoV-2 IgG presence is not enough to protect the second viral infection. According to the fact that reinfection has happened in cases who had developed IgG, the neutralizing potency of the produced antibodies is unknown. There is also another consideration about new viral mutations which are not recognized by existing immunity. Although the distinct between the re-infection and reactivation of SARS-CoV-2 is challenging, these results might be clues of SARS-CoV-2 potency to circulate continuously between the populations despite herd immunity either with natural infection or vaccination. We can conclude that asymptomatic patients in the second infection could strongly impose a threat to the society. Therefore, PCR test must be considered to determine the second incidence which is really critical in term of the patients, clinical investigate, vaccination study, and viral transmission.
References
Epaulard O, Adam L, Poux C et al (2014) Macrophage- and neutrophil-derived TNF-alpha instructs skin langerhans cells to prime antiviral immune responses. J Immunol (Baltimore, Md : 1950) 193(5):2416–2426
Hung IF-N, Cheng VC-C, Li X et al (2020) SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: a case series. Lancet Infect Dis 20(9):1051–1060
Sewell HF, Agius RM, Stewart M et al (2020) Cellular immune responses to covid-19. BMJ 370:m3018
Changes in antibodies and RNA shedding during early SARS-CoV-2 infection [press release]. News Medicla Life Science, May 22, 2020 2020.
Udugama B, Kadhiresan P, Kozlowski HN et al (2020) Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 14(4):3822–3835
Zhou G, Zhao Q (2020) Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int J Biol Sci 16:1718–1723
Siracusano G, Pastori C, Lopalco L (2020) Humoral Immune Responses in COVID-19 Patients: A Window on the State of the Art. Front Immunol 11(1049)
Catanzaro M, Fagiani F, Racchi M et al (2020) Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct Target Ther 5(1):84
Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short-lasting. Nature Medicine. 2020.
Sofian M, Velayati AA, Banifazl M et al (1946) SARS-CoV-2, a virus with many faces: a series of cases with prolonged persistence of COVID-19 symptoms. Wien Med Wochenschr 2020:1–4
To KK-W, Hung IF-N, Ip JD et al (2020) Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin Infect Dis
García LF (2020) Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front Immunol 11(1441)
Shi Y, Wang Y, Shao C et al (2020) COVID-19 infection: the perspectives on immune responses. Cell Death Differ 27(5):1451–1454
Woelfel R, Corman VM, Guggemos W et al (2020) Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. medRxiv. https://doi.org/10.1101/2020.03.05.20030502
OKBA NMA, Muller MA, Li W et al (2020) SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv. https://doi.org/10.1101/2020.03.18.20038059
Zhao J, Yuan Q, Wang H et al (2019) Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease. Clin Infect Dis : Off Publ Infect Dis Soc Am:2020
Long Q-X, Tang X-J, Shi Q-L et al (2020) Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 26(8):1200–1204
To KK-W, Tsang OT-Y, Leung W-S et al (2020) Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20(5):565–574
Wu J, Liang B, Chen C et al (2020) SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. medRxiv. https://doi.org/10.1101/2020.07.21.20159178
Coppola A, Annunziata A, Carannante N et al (2020) Late Reactivation of SARS-CoV-2: A Case Report. Front Med 7(531)
Luciani M, Bentivegna E, Spuntarelli V, et al Recurrent COVID-19 pneumonia in the course of chemotherapy: Consequence of a weakened immune system? J Med Virol. 2020.
Alizargar J (2020) Risk of reactivation or reinfection of novel coronavirus (COVID-19). J Formos Med Assoc 119(6):1123
Poland GA, Ovsyannikova IG, Kennedy RB SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. The Lancet
The COVID-19 diagnosis and treatment flowchart in Iran (3rd edition). [Internet]. 2020 [cited 18 March, 2020]. Available from: http://treatment.sbmu.ac.ir/index.jsp?siteid=62&fkeyid=&siteid=62&pageid=63989.
Parry J (2020) Covid-19: Hong Kong scientists report first confirmed case of reinfection. BMJ 370:m3340
Lab confirms first case of coronavirus reinfection in the US [Internet]. 2020. Available from: https://www.livescience.com/coronavirus-reinfection-case-confirmed-us.html.
Bonifácio LP, Pereira APS, Araújo DCdAe et al (2020) Are SARS-CoV-2 reinfection and Covid-19 recurrence possible? a case report from Brazil. Rev Soc Bras Med Trop 53
Nachmias V, Fusman R, Mann S et al (2020) The first case of documented Covid-19 reinfection in Israel. IDCases 22:e00970–e0097e
Prado-Vivar BB-W, Monica, Guadalupe, Jose J, Marquez, Sully, Gutierrez, Bernardo, Rojas-Silva, Patricio, Grunauer, Michelle, Trueba, Gabriel, Barragan, Veronica, Cardenas, Paul COVID-19 Re-Infection by a Phylogenetically Distinct SARS-CoV-2 Variant. First Confirmed Event South Am
Iwasaki A What reinfections mean for COVID-19. Lancet Infect Dis
Tillett RL, Sevinsky JR, Hartley PD et al Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis
To KK-W, Hung IF-N, Ip JD et al (2020) COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis
Coronavirus reinfections: three questions scientists are asking [Internet]. Nature. 2020. Available from: https://www.nature.com/articles/d41586-020-02506-y.
Lancman G, Mascarenhas J, Bar-Natan M (2020) Severe COVID-19 virus reactivation following treatment for B cell acute lymphoblastic leukemia. J Hematol Oncol 13(1):131
Bentivegna E, Sentimentale A, Luciani M et al (2020) New IgM seroconversion and positive RT-PCR test after exposure to the virus in recovered COVID-19 patient. J Med Virol
A new Covid variant has been discovered in Brazil — here’s what we know so far [Internet]. CNBC. 2021 [cited 2021/01/19]. Available from: https://www.cnbc.com/2021/01/15/brazil-a-new-covid-variant-has-been-discovered-heres-what-we-know-so-far.html.
Grubaugh ND, Hanage WP, Rasmussen AL (2020) Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell 182(4):794–795
Hu J, He C-L, Gao Q-Z et al (2020) D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. bioRxiv. https://doi.org/10.1101/2020.06.20.161323
Lorenzo-Redondo R, Nam HH, Roberts SC et al (2020) A Unique Clade of SARS-CoV-2 Viruses is Associated with Lower Viral Loads in Patient Upper Airways. medRxiv. https://doi.org/10.1101/2020.05.19.20107144
Acknowledgment
We do appreciate Pasteur Institute of Iran for financial support of this study.
Funding
This work was supported by Pasteur Institute of Iran [grant number 1158].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The study was approved by Pasteur Institute of Iran Ethical committee under IR.PII.REC.1399.009 ethical code.
Conflict of interest
Authors declare no potential conflict of interest that could negatively influence the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Salehi-Vaziri, M., Jalali, T., Farahmand, B. et al. Clinical characteristics of SARS-CoV-2 by re-infection vs. reactivation: a case series from Iran. Eur J Clin Microbiol Infect Dis 40, 1713–1719 (2021). https://doi.org/10.1007/s10096-021-04221-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04221-6