Abstract
Elizabethkingia genus is emerging in hospitals and resistant to multiple antibiotics. The intrinsic imipenem resistance of Elizabethkingia genus is related to two chromosome-encoded metallo-beta-lactamases (MBLs), BlaB and GOB. This study was aimed to investigate the in vitro activity of imipenem, vancomycin, and rifampicin in clinical Elizabethkingia species. The distribution and heterogeneity of MBLs responsible for imipenem resistance were also evaluated. A total of 167 Elizabethkingia isolates from different patients were collected, including E. anophelis (142), E. meningoseptica (11), and E. miricola (14). All isolates were evaluated by the broth microdilution assay, ethylenediaminetetraacetic acid (EDTA) combination disk test, and EDTA-based microdilution test. The characteristics of BlaB and GOB were evaluated in phylogenetic analysis and heterologous expression experiments. Most of the isolates were susceptible to rifampin (94%), whereas none of the isolates were susceptible to imipenem. Vancomycin showed intermediate effectiveness. EDTA could reduce 4 folds or more minimum inhibitory concentrations (MICs) of imipenem in 105 isolates (62.9%). Of the isolates, the amino acid sequences of BlaB and GOB were divided into 22 and 25 different types, respectively. Phylogenetic analysis showed BlaB and GOB are species-specific proteins. Furthermore, GOB and BlaB from E. anophelis showed higher imipenem hydrolysis efficiency than those from the other two species. Rifampicin remained the most active agent in the current study. The mechanism of Elizabethkingia resistance to imipenem primarily stemmed from MBLs but other mechanisms could also exist, which requires further investigation.
Similar content being viewed by others
Introduction
The Elizabethkingia species is an aerobic, non-motile, glucose non-fermenting Gram-negative bacilli (GNBs) frequently detected in the environment and is known to be widely resistant to many classes of antibiotics. In recent years, the incidence of Elizabethkingia infections shows an unexplained growing trend around the world [1]. To date, more than seven species are included in the Elizabethkingia genus [2, 3]. Among them, E. anophelis, E. meningoseptica, and E. miricola were reported to be associated with clinical infections. The mortality rate of the patients with Elizabethkingia infections ranges from 18.2 to 34.2% [4,5,6]. The three clinically related Elizabethkingia species could be identified by the pecific peaks produced by the matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS), 16S ribosomal RNA (rRNA) gene sequencing, and RNA polymerase beta subunit (rpoB) gene sequencing [2, 7,8,9].
Metallo-beta-lactamases (MBLs) are a worldwide concern carbapenemase as they possess activities against carbapenems and all beta-lactam antibiotics (except aztreonam) [10]. MBL activities could be inhibited by chelating agents such as ethylenediaminetetraacetic acid (EDTA) [11, 12]. All Elizabethkingia species carry two chromosome-borne intrinsic MBLs, BlaB (subclass B1) and GOB (subclass B3). Both MBLs are related to the intrinsic resistance to carbapenems [4, 6, 8, 13]. The sequences of BlaB and GOB have revealed heterogeneity, with up to 15 BlaB and 18 GOB alleles identified and registered in the GenBank.
Most infections of E. anophelis and E.miricola were misidentified as E. meningoseptica [14, 15]. Vancomycin and rifampicin have been recommended as the empirical treatment for E. meningoseptica infections in previous research [16, 17]. However, only one study in the literature has evaluated the minimal inhibitory concentrations (MICs) of vancomycin and rifampicin, and the susceptibilities among different Elizabethkingia species [14]. There has been no study to determine the distribution of BlaB and GOB in clinical-related Elizabethkingia species. In this study, we focused on the in vitro activity of imipenem, vancomycin, and rifampicin against the three clinically related Elizabethkingia species. The genotypes and heterogeneity of the genes encoding MBLs were also analyzed.
Material and methods
Bacterial isolates and growth media
From May 2017 to Feb 2018, 167 non-duplicate clinical isolates of the Elizabethkingia species were collected for this study at the Tri-Service General Hospital in Taiwan. The isolates were collected from microbial cultures yielded from 124 respiratory tract specimens, 34 blood samples, 5 abscess samples, 3 urine samples, and 1 pleural fluid sample. The clinical samples were extracted in the microbiology laboratory for routine examination and culture. Cultured pathogens were identified using the Vitek MS matrix-assisted laser desorption ionization–time-of-flight mass spectrometry system (VITEK MS system) (bioMérieux, Mercy l’Etoile, France). The Elizabethkingia species were identified using the Vitek MS system with the IVD 3.0 database (bioMérieux, Mercy l’Etoile, France), sequence analysis of the 16S ribosomal RNA (rRNA) and rpoB gene as described previously [7]. This study also included 10 carbapenem-resistant Klebsiella pneumoniae (CRKP) as a control group for MBL detection. Among them, three were MBL producers (producing IMP-8 type enzyme) and seven were non-producers. Bacteria were grown routinely in the Luria-Bertani (LB) broth (BD Difco, Franklin Lakes, NJ) at 35 °C with shaking. Plasmids were maintained and selected in E. coli DH5alpha and BL21(DE3) using a medium supplemented with kanamycin (Sigma–Aldrich) (25 mg/L).
Antimicrobial susceptibility tests
The minimum inhibitory concentration (MIC) values (mg/L) of imipenem, vancomycin, and rifampicin were evaluated by the broth microdilution assay and the E-test (AB Biodisk, Solna, Sweden). Antimicrobial susceptibility interpretation was based on the Clinical and Laboratory Standards Institute (CLSI) criteria (M100-S27, 2017) [18]. The breakpoint used for imipenem was adapted from that for non-Enterobacteriaceae and those for vancomycin and rifampicin were from Staphylococcus species. Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853 were used as controls. To verify the performance of MBLs, we prepared the Mueller–Hinton broth with and without adding isopropyl -D-thiogalactopyranoside (IPTG) (Sigma–Aldrich) at a 0.025 mM final concentration.
EDTA-imipenem microdilution test
The MICs of imipenem in the presence or absence of EDTA were determined by the broth microdilution assay performed as described previously [11]. The MICs of EDTA alone were more than 1.6 mM for all Elizabethkingia isolates and CRKP isolates. All Elizabethkingia and CRKP isolates grew well in the MHB broth with one-fourth (0.4 mM) of the EDTA MIC. The final concentration of EDTA used in the EMT assay was 0.4 mM. Each isolate was tested in duplicates. In each series, the MICs of imipenem and the imipenem plus EDTA were either identical or differed by a twofold dilution at most. The EDTA effects on the MIC of imipenem were interpreted and reported as good effect when there was a four-fold or more reduction, and otherwise as poor effect [11].
EDTA combination disk test (EDT)
Together with imipenem disks, imipenem/0.1 M EDTA, imipenem/0.2 M, and imipenem/0.5 M EDTA combination disks were employed for detection in the combination disk tests (CDTs) as described previously [12, 19, 20]. In summary, four imipenem (10 μg) disks (BBL, France) were placed on a Mueller–Hinton agar for each tested isolate. Ten milliliters of each 0.1 M, 0.2 M, and 0.5 M EDTA solutions (pH 8.0) was added separately to three of the four imipenem disks. After incubation for 18–20 h at 35 °C, the inhibition zones of the imipenem disks with and without EDTA were compared. An increase of ≧ 4 mm in the zone diameter of the imipenem disk in the presence of 0.1 M EDTA was interpreted as a positive result for MBL production [19]. For those with 0.2 M and 0.5 M EDTA in the imipenem disk, a ≧ 7-mm zone diameter difference was interpreted as a positive result [12, 20].
Phylogenetic analysis of MBL genes
Genomic DNA was isolated using the standard phenol/chloroform protocol suggested in the literature [21]. All oligonucleotides used in the study are shown in Table S1. Primer pairs that can detect BlaB and GOB MBL genes were used for PCR amplification of the target genes. Standard PCR amplifications were performed by Phusion DNA polymerase with proofreading activity (Thermo Scientific). Nucleotide sequencing was performed by the BigDye Terminator Cycle Sequencing kit (Applied Biosystems) according to the manufacturer’s instructions. The nucleotide and protein sequences of BlaB and GOB MBLs were compared and grouped by multiple sequence alignments. All of the novel variants were confirmed by the Sanger sequencing of PCR-amplified segments in at least two independent experiments. Different BlaB and GOB MBL amino acid sequences were aligned using the Clustal W multiple sequence alignments. Phylogenetic trees were constructed using MEGA version X (20). The maximum-likelihood (ML) method, using a Jones–Taylor–Thornton (JTT) distance matrix, was used for the construction of BlaB and GOB MBL protein phylogenetic trees. A bootstrap consensus tree inferred from 1000 replicates was taken to deduce confidence levels for the ML trees.
MBL cloning and expression
The open reading frame (ORF) of BlaB and GOB genes was amplified by PCR using the primers pairs, respectively (see Table S1). The ORF amplimer was digested with NotI and then cloned in pET-41b(+). The expression vectors pET-41b(+) were digested with NdeI and made blunt-ended with the Klenow fragment of DNA polymerase I. The blunt-ended plasmid was then treated with NotI to generate a blunt-end/NotI digested pET-41b(+) plasmid. The PCR products of BlaB and GOB genes were digested by NotI before being ligated into the blunt-end/NotI digested pET-41b(+) plasmid to generate a series of expression plasmid pET-BlaB and pET-GOB. The plasmid was transformed into E. coli BL21(DE3) and selected a LB ager supplemented with kanamycin (Sigma–Aldrich) (25 mg/L).
Pulsed-field gel electrophoresis
Cinical Elizabethkingia species isolates with different BlaB and GOB patterns were genotyped by the pulsed-field gel electrophoresis (PFGE) following the digestion of genomic DNA with ApaI [22]. DNA fragments were separated on 1% SeaKem Gold agarose gel (Lonza Rockland, USA) in 0.5 × tris/borate/ethylenediaminetetraacetic acid buffer with CHEF DRIII (BioRad, Hercules, CA, USA) with 6 V/cm, pulse time from 7 to 35 s for 21 h at 14 °C. The PFGE results of representative isolates were interpreted by a phylogenetic tree analysis (BioNumerics program, Applied Meths). Comparison of the patterns was performed by the unweighted pair group method with arithmetic mean clustering. A similarity coefficient of 85% was selected to define the pulsotype clusters.
Nucleotide sequence accession numbers
The nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under accession numbers MK360026 (BlaB_variant01), MK360027 (BlaB_variant02), MK360028 (BlaB_variant03), MK360029 (BlaB_variant04), MK360030 (BlaB_variant05), MK360031 (BlaB_variant06), MK360032 (GOB_variant01), MK360033 (GOB_variant02), MK360034 (GOB_variant03), MK360035 (GOB_variant04), MK360036 (GOB_variant05), MK360037 (GOB_variant06), MK360038 (GOB_variant07), and MK360039 (GOB_variant08).
Results
Demographic characteristics and microbial cultures
The Elizabethkingia species were isolated from the clinical isolates collected from a total of 167 different patients. There was no temporal or spatial overlap among the infected patients during hospitalization. After sequence analyses, the 167 isolates were identified as E. anophelis (142, 85%), E. meningoseptica (11, 6.6%), and E. miricola (14, 8.4%). The most frequently found source was the respiratory tract. Most of the cultures with Elizabethkingia isolates were polymicrobial (105, 62.9%). The major five co-isolated pathogens were Acinetobacter baumannii complex, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Candida species, and Klebsiella pneumoniae. There was no significant difference in the demographic data and the sources of isolation among the infected patients.
Antimicrobial susceptibilities
The MICs of antibiotics against Elizabethkingia isolates are shown in Table 1. Rifampicin was active against E. anophelis (MIC50/90, 0.5/1 mg/L; 94.4% susceptible), E. meningoseptica (MIC50/90, 0.5/1 mg/L; 100% susceptible), and E. miricola (MIC50/90, 0.75/6 mg/L; 94.4% susceptible). Vancomycin exhibited limited activity against E. anophelis (MIC50/90, 16/16 mg/L; 4.2% resistance), E. meningoseptica (MIC50/90, 8/8 mg/L; 0% resistance), and E. miricola (MIC50/90, 16/128 mg/L; 21.4% resistance). Among the three species, E. meningoseptica showed a lower MIC of vancomycin (MIC = 8 mg/L) than the other two. In contrast, imipenem exhibited ineffective activity in all three species (100% resistant).
Activity of MBLs
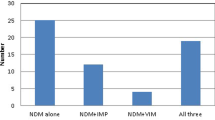
To test whether the activity of MBLs contributed to imipenem resistance, all Elizabethkingia isolates were tested by the EDTA-imipenem microdilution test (EMT) and EDTA combination disk test (ECT). Using EMT experiments, all of the Elizabethkingia isolates exhibited a notable reduction of imipenem MICs in the presence of the mixture of EDTA (Fig. 1). The median MIC value for the isolates was 64 mg/L (range, 16–128 mg/L). This value dropped to 16 mg/L (range, 4–64 mg/L) in the presence of EDTA. When comparing results between different species, the magnitude of reduction ranged from 2 to 8 folds (Table 2). One hundred five (62.9%) of the 167 isolates showed a 4-fold or greater reduction in the MIC of imipenem when EDTA was added. The contribution of MBLs to imipenem resistance was correlated by ECT experiments using different concentrations of EDTA (see Table S2). The inhibitory zones of the imipenem disks increased as the EDTA concentration increased. These data indicate that the MBL activity is involved in imipenem resistance.
Amino acid sequence diversity of MBLs
PCR experiments detected both genes encoding BlaB and GOB MBLs in all clinical isolates. By analyzing the protein sequences, we found that BlaB and GOB contained 248–249 amino acids and 290–292 amino acids, respectively. Amino acid analysis revealed that Elizabethkingia isolates possessed 22 types of BlaB, including 6 novel variants, and 25 types of GOB, including 8 novel variants (see Table S3). The most common combination of MBLs was BlaB-01 (sequence ID: WP_029729112) plus GOB (sequence ID: WP_009089555) (n = 106). Pulsed-field gel electrophoresis (PFGE) was performed on 32 representative isolates with different BlaB and GOB patterns (see Figure S1). All the representative isolates were classified into 3 clusters and the other 12 unique types. The main cluster had 16 isolates containing 3 different species. The phylogenetic tree of BlaB and GOB based on amino acid sequences is shown in Fig. 2. The phylogenetic analysis separated BlaB proteins into four groups. E. anophelis was divided into two groups (groups 1 and 2). E. miricola and E. meningoseptica were clustered into the 2 separate groups. The phylogenetic analysis separated GOB proteins into four distinct isoform groups. E. anophelis was classified into the largest group (group 1). E. miricola was divided into two groups (groups 2 and 3). E. meningoseptica was clustered into group 4. The BlaB and GOB sequences of Elizabethkingia isolates revealed heterogeneity, which is associated with species types.
Phylogenetic inferences of MBL proteins among Elizabethkingia species. An phylogenetic tree was constructed from the BlaB (a) and GOB (b) amino acid sequences by using the maximum likelihood method and JTT matrix-based model [23]. Within the brackets is the sequence ID available in the NCBI database. Evolutionary analyses were conducted in MEGA X [24]
Cloning and heterologous expression of MBLs
To further assess the function of species-specific MBLs, the predominant type of BlaB and GOB genes from the three different species was cloned into pET41b(+) plasmid to construct pET-BlaB and pET-GOB, respectively. The recombinant plasmids were transformed respectively into E. coli BL21(DE3) to obtain the transformed strains. The transformed strains were then tested for imipenem MIC (Table 3). All transformed strains with either pET-BlaB or pET-GOB showed 2–8 folds increases in the MIC of imipenem in comparison with the host strain. Regardless of IPTG stimulation, the BlaB and GOB from E. anophelis showed higher imipenem hydrolysis efficiency than those from E. meningoseptica. The relationship between MBLs and imipenem resistance was verified indirectly by successfully reducing imipenem resistance with EDTA. These results suggested that both MBLs are functionally involved in imipenem resistance in the three clinical-related Elizabethkingia species.
Discussion
E. anophelis was found to be the predominant species in the Elizabethkingia genus in clinical settings [5, 7,8,9, 14]. The older age (≧ 65 years) appears to be a risk factor for clinically related Elizabethkingia species infection as suggested in previous studies [4,5,6, 25]. Senile populations are generally more prone to be hospitalized for infections and thus require antimicrobial treatments. Elizabethkingia species infections are difficult to treat because of intrinsic antimicrobial resistance, and their incidence has recently increased in a global trend [1, 5, 9, 25]. Previous studies showed that the Elizabethkingia species was highly sensitive to piperacillin/tazobactam, doxycycline, minocycline, and rifampin [5, 7, 14]. We found that 94% (157/167) of the Elizabethkingia isolates were sensitive to rifampin as suggested in the literature [14]. Previous studies also indicated that vancomycin was successfully used to cure several patients with bacteremia and meningitis caused by the Elizabethkingia species [6, 16]. Vancomycin, which targets the cell wall peptidoglycan, is bactericidal against Gram-positive bacteria. It is inactive against Gram-negative bacteria due to its inability to penetrate the protective outer membrane [26]. The MIC of vancomycin in the Elizabethkingia species was between 8 and 256 mg/L in our study. This result suggested that a conventional dosage of vancomycin alone might not be sufficient to treat Elizabethkingia infections.
All Elizabethkingia genus harbors two types of chromosomal MBLs (BlaB and GOB) simultaneously [27, 28]. However, another survey in China reported that only 32.4% (55/170) of clinical E. meningoseptica isolates harbors both types of MBLs and 22.4% (38/170) of them harbors a single type of MBL [29]. This might be attributable to the misidentification of the Chryseobacterium species as the Elizabethkingia species in biochemistry tests [9]. Our findings and previous literature have revealed high genetic variability in both MBLs [27, 29]. However, we further point out that the variation of two MBLs is related to the Elizabethkingia species. Chromosome-encoded MBLs may not transmit between Elizabethkingia species and this requires further investigation.
All of our clinical isolates were detected to carry both genes and exhibit high MIC values for imipenem (MIC, 16–128 mg/L). In our EMT experiment, only 62.9% (105/167) of the Elizabethkingia isolates met this criterion. Previous research showed that the EDTA mixture reduced 4-fold or more of the imipenem MICs for MBL producers [11]. These results implied the presence of another imipenem resistant mechanism that was not affected by EDTA. However, in the ECT experiment, the zone size of the imipenem with EDTA was found to be related to the concentration of EDTA added. This phenomenon suggests that high molarity EDTA may induce the membrane permeabilization effect and the zinc ion chelation MBL inhibition effect at the same time [12, 19, 30]. The reductions of outer membrane permeability and carbapenemase hydrolysis are the two main mechanisms of imipenem resistance in Gram-negative bacteria [31]. Accordingly, the high MICs of imipenem against Elizabethkingia isolates may be associated with both MBL hydrolysis and low membrane permeability.
One limitation of this study is that small sample sizes of E. meningoseptica and E. miricola may not represent the antimicrobial susceptibility pattern and MBL gene diversity of these strains in the world. In conclusion, the Elizabethkingia species have emerged in recent years, mainly found in senile patients. Rifampicin remained the most active agent against the Elizabethkingia species. Additionally, our findings suggest that all Elizabethkingia isolates harbor species-specific BlaB and GOB enzymes. To the best of our knowledge, our study is the first one that finds the two MBLs are present in all clinical Elizabethkingia strains. MBL diversity is associated with three clinical Elizabethkingia species and cannot be transferred between species. MBLs are therefore a promising candidate for unique species markers and therapeutic drug development.
References
Choi MH, Kim M, Jeong SJ, Choi JY, Lee IY, Yong TS, Yong D, Jeong SH, Lee K (2019) Risk factors for Elizabethkingia acquisition and clinical characteristics of patients, South Korea. Emerg Infect Dis 25(1):42–51
Kenna DTD, Fuller A, Martin K, Perry C, Pike R, Burns PJ, Narayan O, Wilkinson S, Hill R, Woodford N, Logan JMJ, Turton JF (2018) rpoB gene sequencing highlights the prevalence of an E. miricola cluster over other Elizabethkingia species among UK cystic fibrosis patients. Diagn Microbiol Infect Dis 90(2):109–114
Nicholson AC, Gulvik CA, Whitney AM, Humrighouse BW, Graziano J, Emery B, Bell M, Loparev V, Juieng P, Gartin J, Bizet C, Clermont D, Criscuolo A, Brisse S, McQuiston JR (2018) Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek 111(1):55–72
Lau SK, Chow WN, Foo CH, Curreem SO, Lo GC, Teng JL, Chen JH, Ng RH, Wu AK, Cheung IY, Chau SK, Lung DC, Lee RA, Tse CW, Fung KS, Que TL, Woo PC (2016) Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep 6:26045
Lin JN, Lai CH, Yang CH, Huang YH, Lin HH (2018) Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J Antimicrob Chemother
Figueroa Castro CE, Johnson C, Williams M, VanDerSlik A, Graham MB, Letzer D, Ledeboer N, Buchan BW, Block T, Borlaug G, Munoz-Price LS (2017) Elizabethkingia anophelis: clinical experience of an academic health system in southeastern Wisconsin. Open Forum Infect Dis 4(4):ofx251
Cheng YH, Perng CL, Jian MJ, Cheng YH, Lee SY, Sun JR, Shang HS (2019) Multicentre study evaluating matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically isolated Elizabethkingia species and analysis of antimicrobial susceptibility. Clin Microbiol Infect 25(3):340–345
Chew KL, Cheng B, Lin RTP, Teo JWP (2018) Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol 56(3)
Lin JN, Lai CH, Yang CH, Huang YH, Lin HF, Lin HH (2017) Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci Rep 7(1):13824
Mojica MF, Bonomo RA, Fast W (2016) B1-Metallo-beta-lactamases: where do we stand? Curr Drug Targets 17(9):1029–1050
Migliavacca R, Docquier JD, Mugnaioli C, Amicosante G, Daturi R, Lee K, Rossolini GM, Pagani L (2002) Simple microdilution test for detection of metallo-beta-lactamase production in Pseudomonas aeruginosa. J Clin Microbiol 40(11):4388–4390
Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H (2008) Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother 61(3):548–553
Gonzalez LJ, Vila AJ (2012) Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-beta-lactamase BlaB. Antimicrob Agents Chemother 56(4):1686–1692
Han MS, Kim H, Lee Y, Kim M, Ku NS, Choi JY, Yong D, Jeong SH, Lee K, Chong Y (2017) Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol 55(1):274–280
Janda JM, Lopez DL (2017) Mini review: new pathogen profiles: Elizabethkingia anophelis. Diagn Microbiol Infect Dis 88(2):201–205
Jean SS, Hsieh TC, Ning YZ, Hsueh PR (2017) Role of vancomycin in the treatment of bacteraemia and meningitis caused by Elizabethkingia meningoseptica. Int J Antimicrob Agents 50(4):507–511
Di Pentima MC, Mason EO Jr, Kaplan SL (1998) In vitro antibiotic synergy against Flavobacterium meningosepticum: implications for therapeutic options. Clin Infect Dis 26(5):1169–1176
(CLSI) CaLSI (2017) Performance standards for antimicrobial susceptibility testing, M100-Ed27. Clinical and Laboratory Standards Institute, Wayne
Franklin C, Liolios L, Peleg AY (2006) Phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. J Clin Microbiol 44(9):3139–3144
Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y (2002) Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol 40(10):3798–3801
Syn CK, Swarup S (2000) A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal Biochem 278(1):86–90
Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L (2005) Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43(9):4328–4335
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8(3):275–282
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Perrin A, Larsonneur E, Nicholson AC, Edwards DJ, Gundlach KM, Whitney AM, Gulvik CA, Bell ME, Rendueles O, Cury J, Hugon P, Clermont D, Enouf V, Loparev V, Juieng P, Monson T, Warshauer D, Elbadawi LI, Walters MS, Crist MB, Noble-Wang J, Borlaug G, Rocha EPC, Criscuolo A, Touchon M, Davis JP, Holt KE, McQuiston JR, Brisse S (2017) Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun 8:15483
Munita JM, Arias CA (2016) Mechanisms of antibiotic resistance. Microbiol Spectr 4(2)
Yum JH, Lee EY, Hur SH, Jeong SH, Lee H, Yong D, Chong Y, Lee EW, Nordmann P, Lee K (2010) Genetic diversity of chromosomal metallo-beta-lactamase genes in clinical isolates of Elizabethkingia meningoseptica from Korea. J Microbiol 48(3):358–364
Bellais S, Aubert D, Naas T, Nordmann P (2000) Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. Antimicrob Agents Chemother 44(7):1878–1886
Chen GX, Zhang R, Zhou HW (2006) Heterogeneity of metallo-beta-lactamases in clinical isolates of Chryseobacterium meningosepticum from Hangzhou, China. J Antimicrob Chemother 57(4):750–752
Chu YW, Cheung TK, Ngan JY, Kam KM (2005) EDTA susceptibility leading to false detection of metallo-beta-lactamase in Pseudomonas aeruginosa by Etest and an imipenem-EDTA disk method. Int J Antimicrob Agents 26(4):340–341
Codjoe FS, Donkor ES (2017) Carbapenem resistance: a review. Med Sci (Basel) 6:1
Acknowledgments
The authors thank Ching-Mei Yu and Hung-Sheng Shang for their laboratory work collaboration.
Funding
This study was financially supported by grants from the National Defense Medical Center (MAB-107-014, MAB-107-015, and MAB-107-090), the Taipei City Hospital (TPCH-108-08), and the Ministry of Science and Technology (MOST 108-2320-B-016-009) in Taiwan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed. This work has gone through the ethics committee of our hospital without receiving any kind of objection.
Informed consent
There were no requirements for informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 799 kb)
Rights and permissions
About this article
Cite this article
Chang, TY., Chen, HY., Chou, YC. et al. In vitro activities of imipenem, vancomycin, and rifampicin against clinical Elizabethkingia species producing BlaB and GOB metallo-beta-lactamases. Eur J Clin Microbiol Infect Dis 38, 2045–2052 (2019). https://doi.org/10.1007/s10096-019-03639-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03639-3