Abstract
The significance of the number of coagulase-negative staphylococci (CNS)-positive blood cultures remains obscure in regards to determining true bacteremia versus contamination. The goal of this study was to determine the predictors of real CNS bloodstream infection among intensive care unit (ICU) patients. ICU patients with at least one CNS-positive blood culture were identified from the microbiology database. Biofilm formation was tested by glass tube and microtiter plate assay. mecA gene, ica operon genes (icaA, icaB, icaD), and adhesin genes (aap, bap, atlE, fbe, fnbA) were detected by polymerase chain reaction (PCR). CNS were recovered from 120 septic episodes, 20 of which were true CNS bacteremias, whereas from the remaining 100 episodes, the isolated CNS were characterized as contaminants. The number of positive blood cultures was significantly associated with true CNS bacteremia. Nineteen true bacteremic Staphylococcus epidermidis strains were compared to 38 contaminants. Biofilm synthesis was documented in 37 isolates associated with the presence of the ica operon (p = 0.048). There were 39, 26, 38, 21, and 10 strains positive for the presence of atlE, bap, fbe, aap, and fnbA genes, respectively. Rifampicin resistance, absence of severe sepsis, number of S. epidermidis-positive blood cultures, and absence of the bap gene were independently associated with true S. epidermidis bacteremia as compared to contaminant strains. The number of positive blood cultures is associated with true CNS bacteremia. The presence of adhesin genes may play a role in differentiating true infection from contamination, whereas absence of the bap gene is associated with true S. epidermidis bacteremia.
Similar content being viewed by others
Introduction
Coagulase-negative staphylococci (CNS), especially Staphylococcus epidermidis, are now recognized as an important cause of bloodstream infections (BSIs), especially in patients hospitalized in intensive care units (ICUs) [1]. A few decades back, they were considered as commensal pathogens of the skin [1]. This increase is attributed to the growing number of immunocompromised patients and to increased use of indwelling devices, such as central venous catheters [2].
Even though CNS are commonly isolated from blood cultures, their isolation does not always reflect a true BSI [3–5]. In most cases, they are considered as contaminants. Previous studies tried to identify predictors of true bacteremia, such as the number of positive blood cultures, CNS species, biofilm production, adhesin genes, clonality, etc. [1, 5–8].
Moreover, CNS are important nosocomial pathogens, since they are usually methicillin-resistant, due to the acquisition of mecA gene conferring resistance to all beta-lactams [9]. The fact that CNS are also multidrug-resistant limits our therapeutic options to glycopeptides, daptomycin, and linezolid, whereas resistance to linezolid has already also been observed in our Institution [9, 10].
The goal of the present study was to identify predictors of true CNS BSI among critically septic patients with positive blood cultures. The role of the number of positive blood cultures, the strains’ biofilm formation ability, and their adhesin gene carriage were used in order to differentiate true bacteremia from contamination.
Materials and methods
Patients
This is a retrospective study conducted between December 2009 to February 2012 in the 13-bed ICU of the University Hospital of Patras, Greece.
Patients with at least one positive blood or catheter-tip culture for CNS were identified from the Microbiology Laboratory database. Data regarding demographic characteristics, severity of illness, scores on admission, chronic illnesses, length of hospitalization, surgery, antibiotic usage, and number of catheters inserted were collected from chart reviews and the ICU computerized database (Criticus™, University of Patras, Greece).
Definitions
BSI was defined as systemic symptoms and signs of sepsis with at least two positive blood cultures (7 days apart) with the phenotypically same CNS strain [11]. Catheter-related BSI (CR-BSI) was defined when the central venous catheter tip grew over 15 colony-forming units of the phenotypically same strain as the blood culture isolate by the rolling plate method. Positive blood or catheter-tip cultures not meeting the aforementioned criteria were considered as contaminants.
CNS isolates were identified to the species level by Gram stain, catalase production, and the VITEK 2 Advanced Expert System (bioMérieux, Marcy l’Etoile, France). Antibiotic susceptibility testing was performed by the disk diffusion method against cefoxitin (FOX), erythromycin (E), tetracycline (TET), rifampicin (RIF), clindamycin (CC), kanamycin (KAN), gentamicin (GM), ciprofloxacin (CIP), fusidic acid (FA), and sulfamethoxazole/trimethoprim (SXT) [12]. A gradient method (Etest, bioMérieux) was used to determine the minimum inhibitory concentration (MIC) of oxacillin (OX), vancomycin (VA), teicoplanin (TEC), linezolid (LNZ), and daptomycin (DAP). The results were interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [12].
Biofilm synthesis and presence of virulence factors
Biofilm formation and presence of ica operon, as well as genes encoding adhesins, were determined in 19 infecting and 38 contaminant S. epidermidis isolates (randomly selected from patients with non-CNS sepsis) in order to perform a 1:2 case–control study. Biofilm formation was tested by a qualitative (glass tube assay) and quantitative microtiter plate assay, as previously described [13, 14]. Reference S. epidermidis ATCC35984 (RP62A) and ATCC12228 strains were used in both assays as positive and negative controls, respectively. The detection of mecA gene, genes of ica operon (icaA, icaB, icaD), and the adhesin genes aap, bap, atlE, fbe, and fnbA was performed by polymerase chain reaction (PCR) [15–19].
Statistical analysis
Statistical analyses were performed with SPSS version 19.0 (SPSS, Chicago, IL, USA) software. Continuous variables were assessed with the Mann–Whitney U-test and categorical variables were analyzed by using Fisher’s exact test or the Chi-squared test, as appropriate. Monthly isolation trend during the study period was assessed using Spearman’s correlation analysis. Factors contributing to multicollinearity were excluded from the multivariate analysis. Backward stepwise multiple logistic regression analysis used all variables with a p < 0.1 from the univariate analysis. Odds ratios (ORs) and 95 % confidence intervals (CIs) were calculated to evaluate the strength of any association. All statistical tests were two-tailed and p < 0.05 was considered statistically significant.
Results
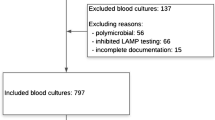
Among the 804 patients admitted to the ICU during the study period, a total of 221 CNS were isolated during 120 septic episodes from 113 patients (2.9 isolations per 100 patient-days) (Fig. 1). The majority were S. epidermidis (167), followed by S. hominis (23), S. haemolyticus (22), and S. capitis (9). Among the 120 septic episodes, 20 (0.3 infections per 100 patient-days) were caused by CNS BSI (19 S. epidermidis and one S. hominis), whereas from the remaining 100 episodes, a total of 176 CNS (82 S. epidermidis and 94 non-epidermidis) were isolated and characterized as contaminants. The contamination rate was 79.6 %. Non-CNS septic episodes were provoked by 43 Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae (28 primary BSI, 12 CR-BSI, two intra-abdominal infections, and one ventilator-associated pneumonia, VAP), 26 Acinetobacter baumannii (24 primary BSI and two meningitis), 18 Pseudomonas aeruginosa (12 VAP and six primary BSI), four Enterococcus faecium (two intra-abdominal infections and two primary BSI), five Escherichia coli (five urinary-tract infections), two Candida albicans (two primary BSI), one Proteus mirabilis (one urinary-tract infection), and one S. aureus (one VAP). Table 1 depicts the univariate analysis of predictors for true CNS BSI as compared to patients with CNS contamination. Multivariate analysis revealed that the number of positive blood cultures for CNS (p < 0.001; OR 28.0; 95 % CI 6.4–122.7) was independently associated with CNS BSI, whereas patients with non-CNS infection had higher Simplified Acute Physiology Score (SAPS) II score on the day of CNS isolation (p = 0.025; OR 0.882; 95 % CI 0.790–0.985) and higher rates of more than one CNS species isolation (p < 0.002; OR 0.001; 95 % CI 0.001–0.089).
Figure 2 depicts the monthly isolation of CNS strains and Fig. 3 shows the resistance patterns of CNS. The monthly isolation of CNS increased during the study period (r 0.407, p = 0.035). Most of the S. epidermidis (155/167, 92.8 %) and all non-S. epidermidis CNS (54/54) were mecA-positive.
Resistant patterns of CNS (167 Staphylococcus epidermidis and 54 non-S. epidermidis) isolated from ICU patients. Susceptibility to vancomycin, linezolid, and daptomycin was assessed with the Etest. FOX cefoxitin; KAN kanamycin; TET tetracycline; RIF rifampicin; GM gentamicin; E erythromycin; CC clindamycin; FA fusidic acid; SXT sulfamethoxazole/trimethoprim; CIP ciprofloxacin; VA vancomycin; LNZ linezolid; DAP daptomycin
S. epidermidis BSI strains (19) were compared to 38 random contaminants in order to determine the role of biofilm formation and presence of adhesin genes as predictors for true CNS BSI (Fig. 1). Of all the tested isolates, biofilm synthesis was observed in 24 (42.1 %) with the glass tube method and in an additional 13 isolates (37, 64.9 %) by the microtiter plate assay, whereas 39 (68.4 %), 38 (66.7 %), 26 (45.6 %), 21 (36.8 %), and 10 (17.5 %) were positive for the presence of atlE, fbe, bap, aap, and fnbA genes, respectively. Biofilm formation was associated with the presence of ica operon (positive PCR for one or more ica gene) (73.0 % vs. 45.0 %; p = 0.048). Table 2 shows the univariate analysis among BSI-causing strains and the contaminants. Rifampicin resistance (p = 0.019; OR 118.3; 95 % CI 2.2–6468.8), absence of severe sepsis or septic shock (p = 0.039; OR 22.3; 95 % CI 1.2–424.1), number of S. epidermidis-positive blood cultures (p = 0.006; OR 24.8; 95 % CI 2.5–344.9), and absence of the bap gene (p = 0.048; OR 17.3; 95 % CI 1.0–292.8) were independently associated with S. epidermidis BSI.
Discussion
CNS are commonly isolated from critically ill patients [1, 5]. The high discrepancy among CNS isolation and infection (2.9 isolations vs. 0.3 infections per 100 patient-days) can be attributed to the fact that CNS are often contaminants as normal skin commensals. Previous studies show that CNS isolates represent contamination (46.4–86.8 %) [3, 4]. The high CNS contamination rate (79.6 %) in the present study may be explained by the low nurse-to-patient ratio in Greek ICUs. This fact increases nurse workload, leading to such an effect due to the overlooking of aseptic standards, as previously reported [5].
Differentiating infection from contamination is difficult, since CNS are part of epidermis normal flora [4]. According to established criteria, two positive blood cultures for CNS are needed in order to determine a BSI infection [11]. In our study, the number of positive blood cultures was independently associated with BSI infection, since all CNS infections had at least two sets of positive blood cultures, as previously shown [3]. On the contrary, other studies have described true CNS infection with only one positive blood culture, even though they did not fulfill the Centers for Disease Control and Prevention (CDC) criteria [3, 20]. In our study, 23 episodes (24.7 %) characterized as contamination had received antibiotic treatment against Gram-positive pathogens, underlying the importance of determining true BSI, since contamination leads not only to unnecessary antibiotic consumption, but also to additional testing and consultation.
The majority of our isolates were methicillin-resistant, a finding consistent with recent studies [9, 19, 21]. Most isolates were also multidrug-resistant, since resistance to aminoglycosides, erythromycin, fusidic acid, co-trimoxazole, and ciprofloxacin surpassed 50 %, limiting our therapeutic armamentarium against these pathogens [9, 22]. On the contrary, the rifampicin resistance rate was low (<30 %) [9, 23]. This fact is reassuring since rifampicin, in vivo and in vitro, shows synergism with other antimicrobial agents, especially in infections associated with foreign bodies, such as central line catheters [24]. Moreover, rifampicin resistance was independently higher among CNS infecting isolates as compared to contaminants. All isolates were susceptible to vancomycin and daptomycin, whereas 55 (24.9 %) isolates (48 S. epidermidis and seven S. capitis) were resistant to linezolid. In a previous study from the same ICU, linezolid resistance among S. epidermidis was due to the presence of C2534T and T2504A mutations in the 23S rRNA gene and to G2576T mutation among S. capitis [10]. As shown, their dissemination was promoted by linezolid administration and mainly to the presence of nearby beds colonized patients [10].
S. epidermidis is more commonly associated with true bacteremia as compared to other CNS species [8, 25]. In our study, since 19 (95.0 %) CNS infections were due to S. epidermidis, a comparison with 38 contaminants was performed in order to evaluate possible differences in biofilm production and presence of virulence determinants. Biofilm production remains the most important pathogenicity mechanism among staphylococci, especially CNS [8, 15]. In total, 37 (64.9 %) isolates were biofilm producers, which is comparable to previous reports [8, 19, 26, 27]. Comparison among the qualitative glass tube method and the quantitative microtiter plate assay revealed that the latter is superior in biofilm-producing S. epidermidis identification. More specifically, the former identified 24 strains, whereas the latter identified 37. The superiority of the microtiter plate assay towards the glass tube method was also shown by Stepanović et al. [13]. Biofilm production is also associated with the presence of ica operon (73.0 % vs. 45.0 %; p = 0.048), which is responsible for the production of the polysaccharide intercellular adhesin (PIA) [16, 19, 26, 28]. The fact that ten biofilm-producing isolates did not carry the ica operon indicates that other factors such as other proteinaceous adhesins may play a role in biofilm synthesis [29].
In the present study, even though no difference was found among biofilm producers and non-producers in adhesin genes carriage, the absence of the bap gene was independently associated with S. epidermidis BSI as compared to colonization. The bap gene was found in 25 (43.9 %) isolates. This percentage is significantly higher than previously reported [17, 19, 30]. The protein Bap is commonly isolated from cases of animal mastitis caused by staphylococci and is associated with adherence to surfaces and production of biofilm that is not composed of PIA [31, 32]. Valle et al. [33] reported that Bap promotes the adhesion of S. aureus to different types of epithelial cells, but impairs bacterial internalization to these cells. Even though the aforementioned abilities contribute to the chronicity of staphylococcal infection, they do not allow tissue invasion and development of bacteremia [33]. In our study, bap-positive strains were associated with colonization, whereas absence of the bap gene was associated with true CNS BSI. On the contrary, in previous studies where the presence of the bap gene was not investigated, biofilm synthesis but not the presence of adhesin genes was associated with CR-BSI [6, 7].
As compared to other pathogens, CNS cause more commonly infections of milder severity [2, 34]. This is evident in the present study, since CNS infections were less commonly associated with severe sepsis or septic shock, as well being associated with lower SAPS II score. The majority of severe infections were due to KPC-producing K. pneumoniae and other multidrug-resistant Gram-negative bacteria, such as A. baumannii and P. aeruginosa, which are characterized by increased mortality [35].
The study has several limitations. First, it is a single-center study and antibiotic susceptibility and molecular findings reflect the local epidemiology. Second, even though the 38 contaminant S. epidermidis strains were randomly selected in order to be compared to infecting strains, no statistical difference was observed among them and the 19 patients with infection with regards to demographics, comorbidities, and severity of disease upon admission.
In conclusion, the contamination of blood cultures from CNS remains high and is higher for non-S. epidermidis. The number of positive blood cultures is associated with true CNS BSI. Biofilm production among S. epidermidis was high and was associated with the presence of ica operon. This is the first time that the absence of the bap gene is found to be associated with true S. epidermidis BSI. The presence of this adhesin gene may play a role in differentiating true infection from contamination.
References
Savithri MB, Iyer V, Jones M, Yarwood T, Looke D, Kruger PS, Faogali J, Venkatesh B (2011) Epidemiology and significance of coagulase-negative staphylococci isolated in blood cultures from critically ill adult patients. Crit Care Resusc 13(2):103–107
Patil HV, Patil VC, Ramteerthkar MN, Kulkarni RD (2011) Central venous catheter-related bloodstream infections in the intensive care unit. Indian J Crit Care Med 15(4):213–223
Mirrett S, Weinstein MP, Reimer LG, Wilson ML, Reller LB (2001) Relevance of the number of positive bottles in determining clinical significance of coagulase-negative staphylococci in blood cultures. J Clin Microbiol 39(9):3279–3281
Rahkonen M, Luttinen S, Koskela M, Hautala T (2012) True bacteremias caused by coagulase negative Staphylococcus are difficult to distinguish from blood culture contaminants. Eur J Clin Microbiol Infect Dis 31(10):2639–2644
Souvenir D, Anderson DE Jr, Palpant S, Mroch H, Askin S, Anderson J, Claridge J, Eiland J, Malone C, Garrison MW, Watson P, Campbell DM (1998) Blood cultures positive for coagulase-negative staphylococci: antisepsis, pseudobacteremia, and therapy of patients. J Clin Microbiol 36(7):1923–1926
Mekni MA, Bouchami O, Achour W, Ben Hassen A (2012) Strong biofilm production but not adhesion virulence factors can discriminate between invasive and commensal Staphylococcus epidermidis strains. APMIS 120(8):605–611
Rohde H, Kalitzky M, Kröger N, Scherpe S, Horstkotte MA, Knobloch JK, Zander AR, Mack D (2004) Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J Clin Microbiol 42(12):5614–5619
Uyanik MH, Yazgi H, Ozden K, Erdil Z, Ayyildiz A (2014) Comparison of coagulase-negative staphylococci isolated from blood cultures as a true bacteremia agent and contaminant in terms of slime production and methicillin resistance. Eurasian J Med 46(2):115–119
Ma XX, Wang EH, Liu Y, Luo EJ (2011) Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): emergence of teicoplanin-non-susceptible CoNS strains with inducible resistance to vancomycin. J Med Microbiol 60(Pt 11):1661–1668
Papadimitriou-Olivgeris M, Giormezis N, Fligou F, Liakopoulos A, Marangos M, Anastassiou ED, Petinaki E, Filos KS, Spiliopoulou I (2013) Factors influencing linezolid-nonsusceptible coagulase-negative staphylococci dissemination among patients in the intensive care unit: a retrospective cohort study. Chemotherapy 59(6):420–426
Horan TC, Andrus M, Dudeck MA (2008) CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 36(5):309–332
European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2015) Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0. EUCAST. Available online at: http://www.eucast.org. Accessed 28 August 2015
Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Cirković I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115(8):891–899
Christensen GD, Simpson WA, Bisno AL, Beachey EH (1982) Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun 37(1):318–326
Cafiso V, Bertuccio T, Santagati M, Campanile F, Amicosante G, Perilli MG, Selan L, Artini M, Nicoletti G, Stefani S (2004) Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect 10(12):1081–1088
Petrelli D, Zampaloni C, D’Ercole S, Prenna M, Ballarini P, Ripa S, Vitali LA (2006) Analysis of different genetic traits and their association with biofilm formation in Staphylococcus epidermidis isolates from central venous catheter infections. Eur J Clin Microbiol Infect Dis 25(12):773–781
Potter A, Ceotto H, Giambiagi-Demarval M, dos Santos KR, Nes IF, Bastos Mdo C (2009) The gene bap, involved in biofilm production, is present in Staphylococcus spp. strains from nosocomial infections. J Microbiol 47(3):319–326
Gomes AR, Vinga S, Zavolan M, de Lencastre H (2005) Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 49(1):366–379
Giormezis N, Kolonitsiou F, Foka A, Drougka E, Liakopoulos A, Makri A, Papanastasiou AD, Vogiatzi A, Dimitriou G, Marangos M, Christofidou M, Anastassiou ED, Petinaki E, Spiliopoulou I (2014) Coagulase-negative staphylococcal bloodstream and prosthetic-device-associated infections: the role of biofilm formation and distribution of adhesin and toxin genes. J Med Microbiol 63(Pt 11):1500–1508
Beekmann SE, Diekema DJ, Doern GV (2005) Determining the clinical significance of coagulase-negative staphylococci isolated from blood cultures. Infect Control Hosp Epidemiol 26(6):559–566
Brzychczy-Wloch M, Borszewska-Kornacka M, Gulczynska E, Wojkowska-Mach J, Sulik M, Grzebyk M, Luchter M, Heczko PB, Bulanda M (2013) Prevalence of antibiotic resistance in multi-drug resistant coagulase-negative staphylococci isolated from invasive infection in very low birth weight neonates in two Polish NICUs. Ann Clin Microbiol Antimicrob 12:41
Tashiro M, Izumikawa K, Ashizawa N, Narukawa M, Yamamoto Y (2015) Clinical significance of methicillin-resistant coagulase-negative staphylococci obtained from sterile specimens. Diagn Microbiol Infect Dis 81(1):71–75
Shah MU, Akram MF, Usman J, Kaleem F (2014) Incidence and susceptibility pattern of methicillin resistant coagulase-negative staphylococci isolated from a tertiary care hospital of Pakistan. Jundishapur J Microbiol 7(1):e8590
Olson ME, Slater SR, Rupp ME, Fey PD (2010) Rifampicin enhances activity of daptomycin and vancomycin against both a polysaccharide intercellular adhesin (PIA)-dependent and -independent Staphylococcus epidermidis biofilm. J Antimicrob Chemother 65(10):2164–2171
Finkelstein R, Fusman R, Oren I, Kassis I, Hashman N (2002) Clinical and epidemiologic significance of coagulase-negative staphylococci bacteremia in a tertiary care university Israeli hospital. Am J Infect Control 30(1):21–25
Ninin E, Caroff N, Espaze E, Maraillac J, Lepelletier D, Milpied N, Richet H (2006) Assessment of ica operon carriage and biofilm production in Staphylococcus epidermidis isolates causing bacteraemia in bone marrow transplant recipients. Clin Microbiol Infect 12(5):446–452
Patel JD, Colton E, Ebert M, Anderson JM (2012) Gene expression during S. epidermidis biofilm formation on biomaterials. J Biomed Mater Res A 100(11):2863–2869
Büttner H, Mack D, Rohde H (2015) Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front Cell Infect Microbiol 5:14
O’Gara JP (2007) ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 270(2):179–188
Darwish SF, Asfour HA (2013) Investigation of biofilm forming ability in Staphylococci causing bovine mastitis using phenotypic and genotypic assays. ScientificWorldJournal 2013:378492
Salaberry SR, Saidenberg AB, Zuniga E, Melville PA, Santos FG, Guimarães EC, Gregori F, Benites NR (2015) Virulence factors genes of Staphylococcus spp. isolated from caprine subclinical mastitis. Microb Pathog 85:35–39
Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR (2001) Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183(9):2888–2896
Valle J, Latasa C, Gil C, Toledo-Arana A, Solano C, Penadés JR, Lasa I (2012) Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS Pathog 8(8):e1002843
Sitges-Serra A, Puig P, Jaurrieta E, Garau J, Alastrue A, Sitges-Creus A (1980) Catheter sepsis due to Staphylococcus epidermidis during parenteral nutrition. Surg Gynecol Obstet 151(4):481–483
Papadimitriou-Olivgeris M, Marangos M, Christofidou M, Fligou F, Bartzavali C, Panteli ES, Vamvakopoulou S, Filos KS, Anastassiou ED (2014) Risk factors for infection and predictors of mortality among patients with KPC-producing Klebsiella pneumoniae bloodstream infections in the intensive care unit. Scand J Infect Dis 46(9):642–648
Acknowledgments
This study was supported partially by funds from the Department of Microbiology, School of Medicine, University of Patras, and by the funding of the National Staphylococcal Reference Laboratory, Greece, under the scientific responsibility of I.S. and E.D.A. (Grant C954, Hellenic Center for Disease Control and Prevention, HCDCP/KEELPNO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study was carried out under the Hospital Surveillance Programme for multi-drug resistant bacterial colonization of patients at risk and HCWs, and was approved by the University Hospital Ethics Committee, which waived the need for informed consent (HEC no.: 571).
Rights and permissions
About this article
Cite this article
Papadimitriou-Olivgeri, I., Giormezis, N., Papadimitriou-Olivgeris, M. et al. Number of positive blood cultures, biofilm formation, and adhesin genes in differentiating true coagulase-negative staphylococci bacteremia from contamination. Eur J Clin Microbiol Infect Dis 35, 57–66 (2016). https://doi.org/10.1007/s10096-015-2506-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2506-7