Abstract
The diversity and complexity of bioinformatics resources presents significant challenges to their localisation, deployment and use, creating a need for reliable systems that address these issues. Meanwhile, users demand increasingly usable and integrated ways to access and analyse data, especially within convenient, integrated “workbench” environments. Resource descriptions are the core element of registry and workbench systems, which are used to both help the user find and comprehend available software tools, data resources, and Web Services, and to localise, execute and combine them. The descriptions are, however, hard and expensive to create and maintain, because they are volatile and require an exhaustive knowledge of the described resource, its applicability to biological research, and the data model and syntax used to describe it. We present here the Workbench Integration Enabler, a software component that will ease the integration of bioinformatics resources in a workbench environment, using their description provided by the existing ELIXIR Tools and Data Services Registry.
Similar content being viewed by others
1 Introduction
Ongoing advances in bioinformatics have produced a vast and ever-increasing number of computational methods and biological databases, available in multiple forms, such as downloadable software or data and remote services for data analysis, query, and retrieval. The diversity and complexity presents significant challenges to resource description, localisation and deployment. Meanwhile, users demand increasingly convenient and usable ways to access and analyse data, especially within environments that integrate resources or handle workflow. We propose a novel approach to integration of existing resources in such environments, that reuses resource descriptions extracted from the ELIXIR Tools and Data Services Registry [27, 28], hereon referred to as “ELIXIR registry”.
Registries address the question of resource discovery, i.e. finding and understanding relevant resources, by collating resource descriptions into a searchable catalogue. Examples of registries within bioinformatics include the EMBRACE Web Service Registry [20], BioCatalogue [1], and AppDB [9]. Other systems, such as BioMoby [29] and Soaplab [24, 25], were developed to enable both the description and the execution of services, using a Web-based interface. Such registries face significant challenges. Solutions based on Web-service technologies do not always scale to the large data volumes required for high-throughput omics analyses. Furthermore, centralised registry efforts have tended to deteriorate in the long-term, and are only fulfilling the discovery purpose, where they have not been coupled to environments for accessing the resources. ELIXIR [8], a European infrastructure for biological information, is constructing the Tools and Data Services Registry for bioinformatics resources from around the world. The registry is being built through collaboration with the key resources providers, upon a federated curation model which supports resource providers in the curation of their own resources. This model decentralises the curation burden and should not only lead to a registry that is more durable, but one that is of higher quality because it leverages the knowledge of the resource providers. In this article, we are outlining the vision of coupling the ELIXIR registry to workbench environments to avoid duplication of curation efforts and maximise utility for users.
While registries address the question of resource discovery, the usage of the tools often remains difficult, because their configuration may be complex and their execution rely on command-line or programmatic interfaces, which are not always transparent to the user. To enhance accessibility, usability, and combining tools, workbenches enable tools execution using graphical, often Web-based, user interfaces. Most of these systems (e.g. Mobyle [17, 18], Galaxy [2, 10], Bio-jETI [15, 16], GenePattern [21], UGENE [19], Geneious [13], and Taverna [14, 30]) rely on detailed tool descriptions in a plugin-based architecture that automatically generates the user interface, invokes the tool, and displays the results in a homogeneous environment. Additionally, workbenches use the tool descriptions for other essential functions, such as searching for and explaining tools, and workflow composition. Thus, there are significant functional and conceptual overlaps between registries and workbenches, which are not reflected in the existing, uncoupled registry and workbench implementations.
The registration or integration of resources, whether in registry or workbench systems, relies on resource descriptions. The structure of such documents is described in detail in Sect. 2. The format of such plugin documents is usually complex and highly specific to the target system. Furthermore, because of the inherent complexity of tools, the descriptions can be difficult to create and maintain, especially by a registry or workbench curator who is not necessarily as familiar with a tool as the person who developed it. This leads to multiple recurring problems that have been addressed in various ways, as described in Sect. 4. In Sect. 5, we describe the new and complementary approach to this problem that we are currently developing; the semi-automatic generation of workbench integration components from the descriptions of resources registered in the ELIXIR Tools and Data Services Registry. This Workbench Integration Enabler will be a software component that eases the integration of bioinformatics resources in workbench or workflow environment such as Mobyle, Galaxy and Taverna, using their description provided by the existing ELIXIR Tools and Data Services Registry.
2 Resource descriptions for registries and workbench systems
Registries and workbench systems both rely on a data model that enables the description of resources they integrate. However, because their functionalities are different, the information stored about the resources in both types of systems overlaps, but is not identical.
2.1 Resource descriptions for registries
The resource description within a registry has to support the use cases related to resource discovery, which include:
-
find a resource by various means, for example, based on the operation that needs to be performed, its inputs and outputs, by the name of the resource or its author, by searching its description, or by the type of interface that is required
-
verify the relevance of a selected resource by reading its description, the publication it refers to, the available documentation, by comparing it to existing offerings, etc.
-
access the resource, which might for example be a Web service or a downloadable and installable package
-
cite the resource in a publication.
Based on these use cases, a description for a tool in a registry might include:
-
the name of the tool
-
a URL to access directly or to download the tool
-
a short and human-readable description
-
the list of its authors, and the list of the publications describing the tool
-
the descriptions of the specific operations that are implemented and the types of data they process and produce, in both human and machine-understandable forms.
2.2 Resource descriptions for workbenches
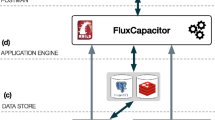
In contrast with a registry, tool descriptions for a workbench must not only support its users in finding and understanding the tools, but also handle their integration into a homogeneous environment to facilitate their execution. Tools can only be integrated into a workbench if they have an execution interface which can be programmatically accessed, such as an API (Application Programming Interface) or a command-line interface. For instance, the Mobyle workbench allows the execution of command-line programs. The transformation of a user request for the execution of a program, i.e. the transformation from a set of inputs into an executable command, and the capture of the results, as well as the generation of the user interface, are based on the tool descriptions which contain a program-specific adapter code (see Fig. 1). In addition to the functionality for execution, Mobyle uses the tool description to facilitate:
-
search, by enabling users to select relevant tools based on their classification or human-readable description fields
-
combining tools, by filtering only the ones that can be chained together either interactively or automatically for successive steps in an analysis, based on the compatibility of the types of data and formats the tools consume and produce
-
ancillary tasks like data format detection and conversion, by using external tools declared as format detection/conversion utilities.
3 Comparison of description attributes in the ELIXIR Tools and Data Services Registry and Mobyle workbench
From a conceptual perspective, the data model of a tool description can be divided in three parts:
-
the basic description of the tool is a broad description of what it does and contextual information, such as authors and publications, in both human and machine-interpretable terms. It is mostly used for search purposes.
-
the function describes how to interact with the given tool, by providing a more detailed description of its inputs and outputs, and the operations it can perform.
-
the implementation of the tool enables its automatic execution. In the case of Mobyle, this requires for a command-line tool, a description of how a user’s request is transformed into a command, and how its results are captured once the command has been executed.
The comparison of the data models of the ELIXIR registry and Mobyle (see Table 1) shows that the tool basic description is shared by both types of description. Additionally, the function part is also represented in both, although only the main parameters of a tool (its main inputs and outputs) are usually described in the ELIXIR registry. The only major aspect that is not covered by the registry description is the implementation. Hence, the alignment of the two data models shows there is a solid foundation for their integration.
4 Creation and maintenance of tool descriptions within a workbench
Tool descriptions, used when adding tools either to a registry or a workbench system, are hard and expensive to maintain, because they have a tendency to evolve, and require exhaustive knowledge of both the described tool, and the data model and syntax used for the description. The description of complex tools, that should enable their integration in specific workbench systems, is even the basis of dedicated efforts and projects [6, 7]. For instance, a typical case of providing an interface within Mobyle to a multiple alignment program can involve the creation and description of around 20 different input and output parameters. In general, the cost and complexity of the description process often results in the following problems:
-
the evolution of a tool is not always captured in a timely manner. For instance, a new version of a software may have new input parameters, but this is often not reflected immediately upon deployment, especially if the new inputs are optional. Such a change can happen without notice, because it does not break the existing tool usage, but nevertheless induces a bias in the available interface.
-
the descriptions for workbench environments are not exhaustive with regard to all of the possible options available in the published software, because of bias of the initial intended usage. The time required to describe completely the software can be reduced, by modelling a minimal set of options which are immediately needed, but this limits the potential of the tool or even prevents some niche uses completely.
-
the tool descriptions tend to focus on the execution layer, that enables the execution of the tool but lacks peripheral information that is useful to achieve a greater degree of integration. This is an acute issue for finding tools, their usability (requiring documentation), composition (requiring parameter typing), provenance tracking (requiring a record of settings and their semantics), and attribution (requiring means of accreditation, citation, etc.). Given the importance of these aspects, especially for non-familiar users, this can reduce the utility of such interfaces greatly.
To mitigate the above issues, different approaches have been explored. Some systems provide graphical user interfaces to create and maintain tool descriptions. For instance, CLI-mate [26] provides a Web-based user interface to generate tool descriptions for platforms like Galaxy and MOTEUR [11]. This approach saves the users from learning a complex and platform-specific syntax before describing a tool. Other systems facilitate sharing and reuse of the descriptions. For instance, an advantage of the Galaxy Toolshed [3] is that it allows the Galaxy community to share tool descriptions, and includes a review mechanism to ensure that the shared tools meet a minimal quality standard. Finally, the use of common invocation syntaxes has been useful. For instance, the description of EMBOSS applications uses a grammar [22] to standardize the syntax of the command line. This enabled an almost-effortless integration of command line applications into other environments, such as the wEMBOSS [23] or Jemboss [5] workbenches, or the SoapLab Web Service publication platform. Similarly, other groups of applications that use a common API, such as BioMOBY resources can be easily integrated in client systems such as Taverna or Remora [4].
5 Usage of tool descriptions from ELIXIR Tools and Data Services Registry as templates for creation of tool wrappers for workbenches
We propose here a novel and complementary approach to assist the integration of new services into workbench systems, that reuses the service descriptions from the ELIXIR registry. This approach, starting from the information about a given tool in the registry, maps it to a template that is generated in the target system’s own syntax. The current tool description model in the ELIXIR registry is focused mainly on the tool basic description, and to a certain extent, its function. Therefore, missing information, especially about the tool’s implementation, i.e. how to execute it, must be added, either by extension of the registry’s model or on the workbench side, as appropriate. As an analogy, this resembles the automatic generation of skeleton code from the description of its interface, as offered by programming tools like the Apache Axis Web-services framework.Footnote 1 The Workbench Integration Enabler component (see Fig. 2) will enable the automatic or semi-automatic generation of workbench adapters for bioinformatics workbench or workflow frameworks, such as Mobyle, Galaxy and Taverna.
This approach offers several significant advantages:
-
collaboration between the ELIXIR registry and workbench maintainers—to maintain the information that is required for both the registry and workbenches in one place—will save time and effort, and lead to better tool descriptions and more durable registry and workbench environments. This is especially so, given that ELIXIR is supporting this vital “document once” principle and supporting resource providers in the curation of their own resources.
-
the ELIXIR registry uses the EDAM ontology [12] to provide a controlled vocabulary for the description of scientific topics, software operations, types of data and data formats. By propagating these annotations to the workbench, the end-user will benefit from rich and consistent tool descriptions in both environments. Further, EDAM development will leverage the user communities of both environments ensuring the vocabulary fulfills users needs.
-
updates in the registered tool descriptions can propagate from the ELIXIR registry—via a notification service—to integrators. This will inform integrators about changes and summarise the changes, so that they can be acted on in a timely manner.
-
general information such as the authors and references is emphasized in the ELIXIR registry but tends to be neglected by integrators in the creation of tool descriptions. It will be a valuable complement to the workbench, useful for both tool providers and users.
-
integration of tools with standardized interfaces, such as EMBOSS tools, can be completely automated by merging the technical information provided with the tools (such as the tools descriptions in EMBOSS), with the applicability and attribution information from the ELIXIR Tools and Data Service Registry.
A prototype implementation for the mapping of the schemas of the ELIXIR Registry and Mobyle, and technical transformation of the resource descriptions from those two systems, is under development. Once its implementation is ready for a public release, this tool will be released both as a free and open source tool and as a public website. We welcome collaborations from technical and scientific end-users to develop this project further, who can contact directly the authors.
6 Conclusion
We presented here a novel way to improve the integration of bioinformatics resources in workbench systems, by mapping and translating the resource metadata contained in the ELIXIR registry. This approach can significantly reduce the problems previously cited which hinder the generation and maintenance of resource descriptions, by improving their quality, comprehensiveness and update time. When implemented as a service, it will lower the cost to developers of integrating their resources in key workbench environments, and assist bioinformaticians to build, use and update well documented and reproducible workflows. It will therefore be a practical way to improve resource utility, including interoperability. As new, high priority tools and services are added to the ELIXIR registry, these can be offered as candidates for inclusion in the workbench instances. This will in turn inform and drive the curation of such resources in sufficient detail to support their integration and invocation. We also plan to capitalize on the use of the ELIXIR registry as a reference for both service providers and integrators to facilitate exchanges between these two groups of experts.
References
Bhagat, J., Tanoh, F., Nzuobontane, E., Laurent, T., Orlowski, J., Roos, M., Wolstencroft, K., Aleksejevs, S., Stevens, R., Pettifer, S., et al.: Biocatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 38, W689–W694 (2010). doi:10.1093/nar/gkq394
Blankenberg, D., Kuster, G.V., Coraor, N., Ananda, G., Lazarus, R., Mangan, M., Nekrutenko, A., Taylor, J.: Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. (2010). doi:10.1002/0471142727.mb1910s89
Blankenberg, D., Von Kuster, G., Bouvier, E., Baker, D., Afgan, E., Stoler, N., Taylor, J., Nekrutenko, A., et al.: Dissemination of scientific software with galaxy toolshed. Genome Biol. 15(2), 403 (2014)
Carrere, S., Gouzy, J.: REMORA: a pilot in the ocean of BioMoby web-services. Bioinformatics 22(7), 900–901 (2006)
Carver, T., Bleasby, A.: The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics 19(14), 1837–1843 (2003)
Cock PJA, Grüning BA, Paszkiewicz K, Pritchard L.: Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. Peer J 1:e167 (2013). doi:10.7717/peerj.167
Cock, P.J., Grüning, B.A., Paszkiewicz, K., Pritchard, L.: Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ 1, e167 (2013)
Crosswell, L.C., Thornton, J.M.: ELIXIR: a distributed infrastructure for European biological data. Trends Biotechnol. 30(5), 241–242 (2012)
EGI Application Database (AppDB). https://appdb.egi.eu. Accessed 21 July 2015
Giardine, B., Riemer, C., Hardison, R.C., Burhans, R., Elnitski, L., Shah, P., Zhang, Y., Blankenberg, D., Albert, I., Taylor, J., et al.: Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15(10), 1451–1455 (2005)
Glatard, T., Montagnat, J., Lingrand, D., Pennec, X.: Flexible and efficient workflow deployment of data-intensive applications on grids with MOTEUR. Int. J. High Perform. Comput. Appl. 22(3), 347–360 (2008)
Ison, J., Kalaš, M., Jonassen, I., Bolser, D., Uludag, M., McWilliam, H., Malone, J., Lopez, R., Pettifer, S., Rice, P.: EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics 29(10), 1325–1332 (2013)
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., Drummond, A.: Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12), 1647–1649 (2012). doi:10.1093/bioinformatics/bts199. http://bioinformatics.oxfordjournals.org/content/28/12/1647.abstract
Krabbenhöft, H.N., Möller, S., Bayer, D.: Integrating ARC grid middleware with Taverna workflows. Bioinformatics 24(9), 1221–1222 (2008)
Lamprecht, A.L., Naujokat, S., Margaria, T., Steffen, B.: Semantics-Based Composition of EMBOSS Services with Bio-jETI. In: Proceedings of the Workshop on Semantic Web Applications and Tools for Life Sciences (SWAT4LS). Amsterdam, The Netherlands, November 20, 2009 (2009)
Lamprecht, A.L., Naujokat, S., Margaria, T., Steffen, B.: Semantics-based composition of EMBOSS services. J. Biomed. Semant. 2(S-1), S5 (2011)
Ménager, H., Gopalan, V., Néron, B., Larroudé, S., Maupetit, J., Saladin, A., Tufféry, P., Huyen, Y., Caudron, B.: Bioinformatics applications discovery and composition with the Mobyle suite and Mobylenet. In: Resource Discovery, pp. 11–22. Springer, Berlin (2012)
Néron, B., Ménager, H., Maufrais, C., Joly, N., Maupetit, J., Letort, S., Carrere, S., Tuffery, P., Letondal, C.: Mobyle: a new full web bioinformatics framework. Bioinformatics 25(22), 3005–3011 (2009)
Okonechnikov, K., Golosova, O., Fursov, M., et al.: Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8), 1166–1167 (2012)
Pettifer, S., Ison, J., Kalaš, M., Thorne, D., McDermott, P., Jonassen, I., Liaquat, A., Fernández, J.M., Rodriguez, J.M., Pisano, D.G., et al.: The EMBRACE web service collection. Nucleic Acids Res. 38(suppl 2), W683–W688 (2010)
Reich, M., Liefeld, T., Gould, J., Lerner, J., Tamayo, P., Mesirov, J.P.: Genepattern 2.0. Nat. Genet. 38(5), 500–501 (2006)
Rice, P., Longden, I., Bleasby, A., et al.: EMBOSS: the European molecular biology open software suite. Trends Genet. 16(6), 276–277 (2000)
Sarachu, M., Colet, M.: wEMBOSS: a web interface for EMBOSS. Bioinformatics 21(4), 540–541 (2005)
Senger, M., Rice, P., Bleasby, A., Oinn, T., Uludag, M.: Soaplab2: more reliable sesame door to bioinformatics programs. In: Bioinformatics Open Source Conference, BOSC, vol. 8 (2008)
Senger, M., Rice, P., Oinn, T.: Soaplab-a unified sesame door to analysis tools. In: Proceedings of the UK e-Science All Hands Meeting, vol. 18, pp. 509–513. Citeseer (2003)
Tatum, Z., den Dunnen, J., Laros, J.F.: CLI-mate: an interface generator for command line programs. In: Proceedings of the 4th International Workshop on Semantic Web Applications and Tools for the Life Sciences, pp. 114–115. ACM (2011)
The Danish ELIXIR node. http://elixir-node.cbs.dtu.dk/. Accessed 21 July 2015
The ELIXIR Tools and Data Services Registry. http://elixir-registry.cbs.dtu.dk. Accessed 21 July 2015
Wilkinson, M.D., Links, M.: BioMOBY: an open source biological web services proposal. Brief. Bioinform. 3(4), 331–341 (2002)
Wolstencroft, K., Haines, R., Fellows, D., Williams, A., Withers, D., Owen, S., Soiland-Reyes, S., Dunlop, I., Nenadic, A., Fisher, P., et al.: The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res. 41, W557–W561 (2013). doi:10.1093/nar/gkt328
Acknowledgments
This work was partly funded by ELIXIR, the research infrastructure for life-science data. Hervé Ménager wishes to thank Bertrand Néron, Fabien Mareuil and Olivia Doppelt-Azeroual for their insights on the maintenance of wrappers for workbench systems.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ménager, H., Kalaš, M., Rapacki, K. et al. Using registries to integrate bioinformatics tools and services into workbench environments. Int J Softw Tools Technol Transfer 18, 581–586 (2016). https://doi.org/10.1007/s10009-015-0392-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10009-015-0392-z