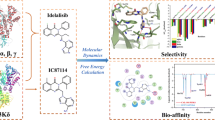
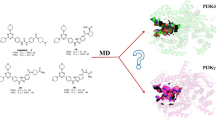
Abstract
Phosphoinositide 3-kinases (PI3Ks) are crucial for cell proliferation, metabolism, motility, and cancer progression. Since the selective PI3Kδ inhibitor, idelalisib, was firstly approved by the FDA in 2014, large numbers of selective PI3Kδ inhibitors have been reported, but the detailed mechanisms of selective inhibition to PI3Kδ for idelalisib or its derivatives have not been well addressed. In this study, 3D-QSAR with COMFA, molecular docking, and molecular dynamic (MD) simulations was used to explore the binding modes between PI3Kδ and idelalisib derivatives. Firstly, a reliable COMFA model (q2 = 0.59, ONC = 8, r2 = 0.966) was built and the contour maps showed that the electrostatic field had more significant contribution to the bioactivities of inhibitors. Secondly, two molecular docking methods including rigid receptor docking (RRD) and induced fit docking (IFD) were employed to predict the docking poses of all the studied inhibitors and revealed the selective binding mechanisms. And then, the results of the MD simulation and the binding free energy decomposition verified that the binding of PI3Kδ/inhibitors was mainly contributed from hydrogen bonding and hydrophobic interactions and some key residues for selective binding were highlighted. Finally, based on the models developed, 14 novel inhibitors were optimized and some showed satisfactory predicted bioactivity. Taken together, the results provided by this study may facilitate the rational design of novel and selective PI3Kδ inhibitors.

.






Similar content being viewed by others
References
Fruman DA, Rommel C (2014) PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 13(2):140–156
Cantley LC (2002) The phosphoinositide 3-kinase pathway. Science 296(5573):1655–1657
Carracedo A, Pandolfi PP (2008) The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene 27(41):5527–5541
Liu P, Cheng H, Roberts TM, Zhao JJ (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8(8):627–644
Courtney KD, Corcoran RB, Engelman JA (2010) The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 28(6):1075–1083
Jiang BH, Liu LZ (2009) PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv. Cancer Res. 102:19–65
Hennessy BT, Smith D, Ram P, Lu Y (2005) Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 4(12):988–1004
Zhu J, Wang M, Cao B, Hou T, Mao X (2014) Targeting the phosphatidylinositol 3-kinase/AKT pathway for the treatment of multiple myeloma. Curr. Med. Chem. 21(27):3173–3187
Setti A, Kumar MJ, Babu KR, Rasagna A (2015) Potency and pharmacokinetics of broad spectrum and isoform-specific p110γ and δ inhibitors in cancers. J. Recept. Signal Transduct. Res. 36(1):26–36
Zhu J, Pan P, Li Y, Wang M, Li D, Cao B, Mao X, Hou T (2014) Theoretical studies on beta and delta isoform-specific binding mechanisms of phosphoinositide 3-kinase inhibitors. Mol. BioSyst. 10(3):454–466
Yuan TL, Cantley LC (2008) PI3K pathway alterations in cancer: variations on a theme. Oncogene 27(41):5497–5510
Jabbour E, Ottmann OG, Deininger M, Hochhaus A (2014) Targeting the phosphoinositide 3-kinase pathway in hematologic malignancies. Haematologica 99(1):7–18
Zhu J, Hou T, Mao X (2015) Discovery of selective phosphatidylinositol 3-kinase inhibitors to treat hematological malignancies. Drug Discov. Today 20(8):988–994
Li T, Wang G (2014) Computer-aided targeting of the PI3K/Akt/mTOR pathway: toxicity reduction and therapeutic opportunities. Int. J. Mol. Sci. 15(10):18856–18891
Li M, Sala V (2018) Phosphoinositide 3-kinase gamma inhibition protects from anthracycline cardiotoxicity and reduces tumor growth. Circulation 138(7):696–711
Vyas P, Vohora D (2016) Phosphoinositide-3-kinases as the novel therapeutic targets for the inflammatory diseases: current and future perspectives. Curr. Drug Targets 18(14):1622–1640
Vangapandu HV, Jain N, Gandhi V (2017) Duvelisib: a phosphoinositide-3 kinase δ/γ inhibitor for chronic lymphocytic leukemia. Expert Opin. Investig. Drugs 26(5):625–632
Lannutti BJ, Meadows SA, Herman SE, Kashishian A (2011) CAL-101, a p110δ selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 117(2):591–594
Sabbah DA, Vennerstrom JL, Zhong HA (2012) Binding selectivity studies of phosphoinositide 3-kinases using free energy calculations. J. Chem. Inf. Model. 52(12):3213–3224
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 3(11):935–949
Sabbah DA, Vennerstrom JL, Zhong HZ (2010) Docking studies on isoform-specific inhibition of phosphoinositide-3-kinases. J. Chem. Inf. Model. 50(10):1887–1898
Shao S, Yu R, Yu Y, Li Y (2014) Dual-inhibitors of STAT5 and STAT3: studies from molecular docking and molecular dynamics simulations. J. Mol. Model. 20(8):2399
Zhang C, Du C, Feng Z, Zhu J, Li Y (2015) Hologram quantitative structure activity relationship, docking, and molecular dynamics studies of inhibitors for CXCR4. Chem. Biol. Drug Des. 85(2):119–136
Iqbal S, Krishnan DA, Gunasekaran K (2018) Identification of potential PKC inhibitors through pharmacophore designing, 3D–QSAR and molecular dynamics simulations targeting Alzheimer’s disease. J Biomol Struct Dyn 36(15):4029–4044
Katari SK, Natarajan P, Swargam S, Kanipakam H, Pasala C (2016) Inhibitor design against JNK1 through e-pharmacophore modeling docking and molecular dynamics simulations. J. Recept. Signal Transduct. Res. 36(6):558–571
Rajamanikandan S, Jeyakanthan J, Srinivasan P (2017) Molecular docking, molecular dynamics simulations, computational screening to design quorum sensing inhibitors targeting LuxP of Vibrio harveyi and its biological evaluation. Appl. Biochem. Biotechnol. 181(1):192–218
Zondagh J, Balakrishnan V, Achilonu I, Dirr HW, Sayed Y (2018) Molecular dynamics and ligand docking of a hinge region variant of South African HIV-1 subtype C protease. J Mol Graph Model 82:1–11
Xu C, Ren Y (2015) Molecular modeling studies of [6,6,5] Tricyclic Fused Oxazolidinones as FXa inhibitors using 3D-QSAR, Topomer CoMFA, molecular docking and molecular dynamics simulations. Bioorg. Med. Chem. Lett. 25(20):4522–4528
Tang HJ, Yang L, Li JH, Chen J (2016) Molecular modelling studies of 3,5-dipyridyl-1,2,4-triazole derivatives as xanthine oxidoreductase inhibitors using 3D-QSAR, Topomer CoMFA, molecular docking and molecular dynamic simulations. J Taiwan Inst Chem E 68:64–73
Aksoydan B, Kantarcioglu I, Erol I, Salmas RE, Durdagi S (2018) Structure-based design of hERG-neutral antihypertensive oxazalone and imidazolone derivatives. J Mol Graph Model 79:103–117
Zhao S, Zhu J, Xu L, Jin J (2017) Theoretical studies on the selective mechanisms of GSK3beta and CDK2 by molecular dynamics simulations and free energy calculations. Chem. Biol. Drug Des. 89(6):846–855
Shen M, Zhou S, Li Y, Li D, Hou T (2013) Theoretical study on the interaction of pyrrolopyrimidine derivatives as LIMK2 inhibitors: insight into structure-based inhibitor design. Mol. BioSyst. 9(10):2435–2446
Xu L, Li Y, Li L, Zhou S, Hou T (2012) Understanding microscopic binding of macrophage migration inhibitory factor with phenolic hydrazones by molecular docking, molecular dynamics simulations and free energy calculations. Mol. BioSyst. 8(9):2260–2273
Ekhteiari Salmas R, Unlu A, Bektas M, Yurtsever M, Mestanoglu M, Durdagi S (2017) Virtual screening of small molecules databases for discovery of novel PARP-1 inhibitors: combination of in silico and in vitro studies. J. Biomol. Struct. Dyn. 35(9):1899–1915
Patel L, Chandrasekhar J, Evarts J (2016) 2,4,6-Triaminopyrimidine as a novel hinge binder in a series of PI3Kδ selective inhibitors. J. Med. Chem. 59(7):3532–3548
Sherman W, Beard H, Farid R (2010) Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 67(1):83–84
Somoza JR, David K (2015) Structural, biochemical, and biophysical characterization of idelalisib binding to phosphoinositide 3-kinase δ. J Bio Chem 290(13):8439–8446
Case DA, Cheatham TE, Darden T, Gohlke H (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26(16):1668–1688
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24(16):1999–2012
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J. Comput. Chem. 25(9):1157–1174
Stewart JPJ (2010) Optimization of parameters for semiempirical methods I. Method. J Comput Chem 10(2):221–264
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the resp model. J. Phys. Chem. 97(40):10269–10280
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Phys. Chem. 98(12):10089–10092
Vincent K, Van Gunsteren W, Hünenberger P (2001) A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22(5):501–508
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Cheminform 32(10):889–897
Hou T, Li Y, Wang W (2011) Prediction of peptides binding to the PKA RIIα subunit using a hierarchical strategy. Bioinformatics 27(13):1814–1821
Sun H, Li Y, Shen M, Tian S, Xu L, Pan P, Guan Y, Hou T (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys. Chem. Chem. Phys. 16(40):22035–22045
Chen F, Liu H, Sun H, Pan P, Li Y, Li D, Hou T (2016) Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. Phys. Chem. Chem. Phys. 18(18):22129–22139
Pan P, Yu H, Liu Q, Kong X, Chen H, Chen J, Liu Q, Li D, Kang Y, Sun H (2017) Combating drug-resistant mutants of anaplastic lymphoma kinase with potent and selective type-I1/2 inhibitors by stabilizing unique DFG-shifted loop conformation. Acs Cent Sci 3(11):1208–1220
Hou T, Li Y, Wang W (2011) Prediction of peptides binding to the PKA RIIalpha subunit using a hierarchical strategy. Bioinformatics 27(13):1814–1821
Sun H, Li Y, Li D, Hou T (2013) Insight into crizotinib resistance mechanisms caused by three mutations in ALK tyrosine kinase using free energy calculation approaches. J. Chem. Inf. Model. 53(9):2376–2389
Sun H, Li Y, Tian S, Xu L, Hou T (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 16(31):16719–16729
Sun H, Duan L, Chen F, Liu H, Wang Z, Pan P, Zhu F (2018) Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys. Chem. Chem. Phys. 20(21):14450–14460
Chen F, Liu H, Sun H, Pan P, Li Y, Li D, Hou T (2016) Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Phys. Chem. Chem. Phys. 18(32):22129–22139
Chen F, Sun H, Wang J, Zhu F, Liu H (2018) Assessing the performance of MM/PBSA and MM/GBSA methods. 8. Predicting binding free energies and poses of protein-RNA complexes. RNA 24(9):1183–1194
Onufriev A, Donald Bashford A, Case DA (2000) Modification of the generalized born model suitable for macromolecules. J. Phys. Chem. B 104(15):3712–3720
Weiser J, Shenkin P, Still W (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J. Comput. Chem. 20(2):217–230
Berndt A, Miller S, Williams O, Le D (2010) The p110δ crystal structure uncovers mechanisms for selectivity and potency of novel PI3K inhibitors. Nat. Chem. Biol. 6(2):117–124
Williams R, Berndt A, Miller S, Hon W, Zhang X (2009) Form and flexibility in phosphoinositide 3-kinases. Biochem. Soc. Trans. 37(4):615–626
Safina BS, Sweeney ZK, Li J, Chan BK (2013) Identification of GNE-293, a potent and selective PI3Kδ inhibitor: navigating in vitro genotoxicity while improving potency and selectivity. Bioorg. Med. Chem. Lett. 23(17):4953–4959
Funding
The study was financially supported by the National Natural Science Foundation of China (No. 21807049, 81803430) and the Fundamental Research Funds for the Central Universities (JUSRP11892).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 2327 kb)
Rights and permissions
About this article
Cite this article
Zhu, J., Ke, K., Xu, L. et al. Theoretical studies on the selectivity mechanisms of PI3Kδ inhibition with marketed idelalisib and its derivatives by 3D-QSAR, molecular docking, and molecular dynamics simulation. J Mol Model 25, 242 (2019). https://doi.org/10.1007/s00894-019-4129-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-019-4129-x