Abstract
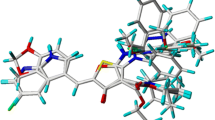
Checkpoint kinase 1 (Chk1), a kind of a serine/threonine protein kinase, plays a significant role in DNA damage-induced checkpoints. Chk1 inhibitors have been demonstrated to abrogate the S and G2 checkpoints and disrupt the DNA repair process, which results in immature mitotic progression, mitotic catastrophe, and cell death. Normal cells remain at the G1 phase via p53 to repair their DNA damages, and are less influenced by the abrogation of S and G2 checkpoint. Therefore, selective inhibitors of Chk1 may be of great therapeutic value in cancer treatment. In this paper, in order to understand the structure-activity relationship of macro-cyclic urea Chk1 inhibitors, a study combined molecular docking and 3D-QSAR modeling was carried out, which resulted in two substructure-based 3D-QSAR models, including the CoMFA model (r2, 0.873; q2, 0.572) and CoMSIA model (r2, 0.897; q2, 0.599). The detailed microscopic structures of Chk1 binding with inhibitors were performed by molecular docking. Two docking based 3D-QSAR models were developed (CoMFA with r2, 0.887; q2, 0.501; CoMSIA with r2, 0.872; q2, 0.520). The contour maps obtained from the 3D-QSAR models in combination with the docked binding structures would be helpful to better understand the structure–activity relationship. All the conclusions drawn from both the 3D-QSAR contour maps and molecular docking were in accordance with the experimental activity dates. The results suggested that the developed models and the obtained CHk1 inhibitor binding structures might be reliable to predict the activity of new inhibitors and reasonable for the future drug design.











Similar content being viewed by others
References
Chen Y, Sanchez Y (2004) Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair 3:1025–1032
Kastan MB, Onyekwere O, Sidransky D et al (1991) Participation of p53 protein in the cellular response to DNA damage. Cancer Res 51:6304–6311
Li Q, Zhu GD (2002) Targeting serine/threonine protein kinase B/Akt and cell-cycle checkpoint kinases for treating cancer. Curr Top Med Chem 2:939–971
Greenblatt MS, Bennett WP, Hollstein M et al (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 54:4855–4878
Yolanda S, Calvin W, Richard ST et al (1997) Conservation of the chk1 checkpoint pathway in mammals: linkage of DNA damage to cdk regulation through Cdc25. Science 277:1497–1501
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Cramer RD, Bunce JD, Patterson DE et al (1988) Crossvalidation, bootstrapping, and partial least squares compared with multiple regression in conventional QSAR studies. Quant Struct Act Relat 7:18–25
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146
Potashman MH, Bready J, Coxon A et al (2007) Design, synthesis, and evaluation of orally active benzimidazoles and benzoxazoles as vascular endothelial growth factor-2 receptor tyrosine kinase inhibitors. J Med Chem 50:4351–4373
Yang GF, Lu HT, Xiong Y et al (2006) Understanding the structure activity and structure-selectivity correlation of cyclic guanine derivatives as phosphodiesterase-5 inhibitors by molecular docking, CoMFA and CoMSIA analyses. Bioorg Med Chem 14:1462–1473
Huang X, Xu L, Luo X et al (2002) Elucidating the inhibiting mode of AHPBA derivatives against HIV-1 protease and building predictive 3D-QSAR models. J Med Chem 45:333–343
Li GQ, Tao ZF, Tong YS (2007) Synthesis and in-vitro biological activity of macro cyclic urea Chk1 inhibitors. Bioorg Med Chem Lett 17:6499–6504
Wang GT, Li GQ (2005) 1-(5-Chloro-2-alkoxyphenyl)-3-(5-cyano- pyrazi-2-yl)ureas as potent and selective inhibitors of Chk1 kinase: synthesis, preliminary SAR, and biological activities. J Med Chem 48:3118–3121
Li GQ, Hasvold LA, Tao ZF (2006) Synthesis and biological evaluation of 1-(2,4,5-trisubstituted phenyl)-3-(5-cyanopyrazin-2-yl)ureas as potent Chk1 kinase inhibitors. Bioorg Med Chem Lett 16:2293–2298
Tao ZF, Wang L, Stewart KD et al (2007) Structure-based design, synthesis, and biological evaluation of potent and selective macrocyclic checkpoint kinase 1 inhibitors. J Med Chem 50:1514–1527
Tao ZF, Chen ZH (2007) Macrocyclic ureas as potent and selective Chk1 inhibitors: an improved synthesis, kinome profiling, structure–activity relationships, and preliminary pharmacokinetics. Bioorg Med Chem Lett 17:6593–6601
Li GQ, Tao ZF, Tong YS (2007) Synthesis and in-vitro biological activity of macrocyclic urea Chk1 inhibitors. Bioorg Med Chem Lett 17:6499–6504
Tao ZF, Wang L, Stewart KD et al (2007) Structure-based design, synthesis, and biological evaluation of potent and selective macrocyclic checkpoint kinase 1 inhibitors. J Med Chem 50:1514–1527
Sybyl Version 7.0 (2004) St. Louis (MO), Tripos Associates Inc. http://www.tripos.com
Clark M, Cramer RD III, Opdenbosch NV (1989) Validation of the general purpose tripos 5.2 force field. J Comput Chem 10:982–1012
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity- a rapid access to atomic charges. Tetrahedron 36:3219–3228
Morris GM, Goodsell DS, Halliday RS et al (1998) Automated docking using a Lamarckian generic algorithm and empirical binding free energy function. J Comput Chem 19:1639–1662
Weiner SJ, Kollman PA, Case DA et al (1984) A new force field for molecular mechanical simulation of nucleic acidsand proteins. J Am Chem Soc 106:765–784
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity– a rapid access to atomic charges. Tetrahedron 36:3219–3228
Mehler EL, Solmajer T (1991) Electrostatic effects in proteins: comparison of dielectric and charge models. Protein Eng 4:903–910
Bush BL, Nachbar RB (1993) Sample-distance partial least squares: PLS optimized for many variables, with application to CoMFA. J Comput Aided Mol Des 7:587–619
Acknowledgments
The authors gratefully acknowledged financial support from the Natural Science Foundation of China (NO. 21071021).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, L., Liu, Y., Hu, S. et al. 3D-QSAR study of Chk1 kinase inhibitors based on docking. J Mol Model 18, 3669–3694 (2012). https://doi.org/10.1007/s00894-012-1363-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1363-x