Abstract
We have performed a multivariate logistic regression analysis to establish a statistical correlation between the structural properties of water molecules in the binding site of a free protein crystal structure, with the probability of observing the water molecules in the same location in the crystal structure of the ligand-complexed form. The temperature B-factor, the solvent-contact surface area, the total hydrogen bond energy and the number of protein–water contacts were found to discriminate between bound and displaceable water molecules in the best regression functions obtained. These functions may be used to identify those bound water molecules that should be included in structure-based drug design and ligand docking algorithms.
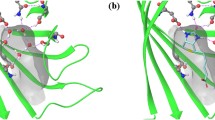
Figure The binding site (thin sticks) of penicillopepsin (3app) with its crystallographically determined water molecules (spheres) and superimposed ligand (in thick sticks, from complexed structure 1ppk). Water molecules sterically displaced by the ligand upon complexation are shown in cyan. Bound water molecules are shown in blue. Displaced water molecules are shown in yellow. Water molecules removed from the analysis due to a lack of hydrogen bonds to the protein are shown in white. WaterScore correctly predicted waters in blue as Probability=1 to remain bound and waters in yellow as Probability<1×10−20 to remain bound.








Similar content being viewed by others
References
Giacovazzo C, Monaco HL, Viterbo D, Scordari F, Gilli G, Zanotti G, Catti M (1992) Fundamentals of crystallography. Oxford University Press, Oxford, pp 583–584
Jeffrey GA (1994) J Mol Struct 322:21–25
Purkiss A, Skoulakis S, Goodfellow JM (2001) Philos Trans R Soc London Ser A 359:1515–1527
Chung E, Henriques D, Renzoni D, Zvelebil M, Bradshaw JM, Waksman G, Robinson CV, Ladbury JE (1998) Struct Folding Design 6:1141–1151
Sanschagrin PC, Kuhn LA (1998) Protein Sci 7:2054–2064
Lemieux RU (1996) Acc Chem Res 29:373–380
Nakasako M (1999) J Mol Biol 289:547–564
Faerman CH, Karplus PA (1995) PROTEINS 23:1–11
Schwabe JWR (1997) Curr Opin Struct Biol 7:126–134
Carrell HL, Glusker JP, Burger V, Manfre F, Tritsch D, Biellmann J-F (1989) Proc Natl Acad Sci USA 86:4440–4444
Baker EL, Hubbard RE (1984) Prog Biophys Molec Biol 44:97–179
Loris R, Langhorst U, De Vos S, Decanniere K, Bouckaert J, Maes D, Transhue TR, Steyaert J (1999) PROTEINS 36:117–134
Loris R, Stas PP, Wyns L (1994) J Biol Chem 269:26722–26733
Poornima CS, Dean PM (1995) J Comput-Aided Mol Des 9:521–531
Poornima CS, Dean PM (1995) J Comput-Aided Mol Des 9:500–512
Poornima CS, Dean PM (1995) J Comput-Aided Mol Des 9:513–520
Feig M, Pettitt BM (1998) Structure 6:1351–1354
Zhang X-J, Matthews BW (1994) Protein Sci 3:1031–1039
Mattos C (2002) Trends Biochem Sci 27:203–208
Esposito L, Vitagliano L, Sica F, Sorrentino G, Zagari A, Mazzarella L (2000) J Mol Biol 297:713–732
Teeter MM (1991) Annu Rev Biophys Chem 20:577–600
Swaminathan CP, Nandi A, Visweswariah SS, Surolia A (1999) J Biol Chem 274:31272–31278
Bhat TN, Bentley GA, Boulot G, Greene MI, Tello D, Dall'Acqua W, Souchon H, Schwarz FP, Mariuzza RA, Poljal RJ (1994) Proc Natl Acad Sci USA 91:1089–1093
Covell DG, Wallqvist A (1997) J Mol Biol 269:281–297
Zhang L, Hermans J (1996) PROTEINS 24:433–438
Helms V, Wade RC (1995) Biophys J 69:810–824
Helms V, Wade RC (1998) PROTEINS 32:381–396
Helms V, Wade RC (1998) J Am Chem Soc 120:2710–2713
Marrone TJ, Briggs JM, McCammon JA (1997) Annu Rev Pharmacol Toxicol 37:71–90
Lam PYS, Jadhav PK, Eyermann CJ, Hodge CN, Ru Y, Bacheler LT, Meek JL, Otto MJ, Rayner MM, Wong YN, Chang CH, Weber PC, Jackson DA, Sharpe, TR, Ericksonviitanen S (1994) Science 263:380–384
Mikol V, Papageorgiou C, Borer X (1995) J Med Chem 38:3361–3367
Palomer A, Pérez JJ, Navea S, Llorens O, Pascual J, García Ll, Mauleón D (2000) J Med Chem 43:2280–2284
Cherbavaz DB, Lee ME, Stroud RM, Koschl DE (2000) J Mol Biol 295:377–385
Finley JB, Atigadda VR, Duarte F, Zhao JJ, Brouillette WJ, Air GM, Luo M (1999) J Mol Biol 293:1107–1119
Ehrlich L, Reckzo M, Wade RC (1998) Protein Eng 11:11–19
Raymer ML, Sanschagrin PC, Punch WF, Venkataram S, Goodman ED, Kuhn L (1997) J Mol Biol 265:445–464
Carugo O (1999) Protein Eng 12:1021–1024
Carugo O, Argos P (1998) PROTEINS 31:201–213
Carugo O, Bordo D (1999) Acta Crystallogr Sect D 55:479–483
Rarey M, Kramer B, Lengauer T (1999) PROTEINS 34:17–28
Pastor M, Cruciani G, Watson KA (1997) J Med Chem 40:4089–4102
Shoichet BK, Leach AR, Kuntz ID (1999) PROTEINS 34:4–16
Mancera RL (2002) J Comp-Aided Mol Des 16:479–499
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235–242
Vriend G (1990) J Mol Graph 8:52–56
Hooft RWW, Sander C, Vriend G (1996) PROTEINS 26:363–376
Hubbard SJ, Argos P (1995) Protein Eng 8:1011–1015
Lee B, Richards FM (1971) J Mol Biol 55:379–400
Matlab 5.0 (1999) The Math Works,
Menard SM (1995) Applied logistic regression analysis in series. In: Lewis-Beck MS (ed) Quantitative applications in the social sciences. Sage, Thousand Oaks, Calif.
Agresti A (1996) An introduction to categorical data analysis, Wiley series in probability and statistics, applied probability and statistics. Wiley, New York
Rice JA (1995) Mathematical statistics and data analysis, 2nd edn. Duxbury Press, Belmont, Calif.
Holtsberg A (1994) http://www.mathtools.net
Acknowledgements
ATGS would like to thank Consejo Nacional de Ciencia y Tecnología (CONACyT, México) for the award of a postgraduate scholarship and the CVCP of the Universities of the UK for an Overseas Research Scheme award. RLM is also a Research Fellow of Hughes Hall, Cambridge. We also thank Mr. Benjamin Carrington for his valuable help in the production of some of the figures, Dr. Per Kållblad for help and discussion on PC analysis, and Miss Eva-Liina Asu for proof-reading a draft of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
We provide a brief outline of multivariate logistic regression analysis. [50, 51, 52] For a binary dependent variable Y that can take values of either 0 or 1, its mean is the proportion of cases of the higher value (1), and the predicted value of the dependent variable (the conditional mean, given the value of the independent variable X and the assumption that Y and X are linearly related) can be interpreted as the predicted probability that an observation falls into such higher value. By definition, the predicted probability lies between 0 and 1. The general shape of the relationship between the probability P(Y=1) and the independent variable X is that of an "S curve", as depicted in Fig. 8.
Instead of predicting the arbitrary value associated with the dependent variable Y, it may be useful to predict the probability that a given observation (as defined by a set of independent variables) will be classified into one of the two values of the dependent variable. Naturally, if we know P(Y=1), we immediately also know the probability of P(Y=0) as P(Y=0)=1−P(Y=1).
If the probability that Y=1 is modeled as P(Y=1)=α+βX, its predicted values may be less than 0 or greater than 1. The first step to avoid this is to replace the probability that Y=1 with the odds that Y=1. The odds that Y=1, written Odds(Y=1), is the ratio of the probability that Y=1 to the probability that Y≠1. Odds(Y=1) is then equal to P(Y=1)/[1−P(Y=1)]. Unlike P(Y=1), the odds has no fixed maximum value, but like the probability, it has a minimum value of 0.
One further transformation of the odds produces a variable that varies, in principle, from negative infinity to positive infinity. The natural logarithm of the odds, ln{P(Y=1)/[1−P(Y=1)]}, is called the logit of Y, and is written logit(Y). This function becomes negative and increasingly large as the odds decrease from 1 to 0, and becomes positive and increasingly large as the odds increase from 1 to infinity. By using the natural logarithm of the odds that Y=1 as the dependent variable, one no longer has the problem that the estimated probability may exceed the maximum or minimum possible values for the probability (see Fig. 8). The equation for the relationship between the dependent variable and a number of independent variables can be then expressed as
Calculating back the odds as Odds(Y=1)=exp[logit(Y)] gives us
A change of unit in X i multiplies the odds by exp(β). The odds can be converted back to the probability that Y=1 by the formula P(Y=1)=Odds(Y=1)/[1+Odds(Y=1)], producing the equation
For any given case, logit(Y)=± ∞. This ensures that the probabilities estimated will not be less than 0 or greater than 1. Because the linear form of the model (Eq. 4) can have infinitely large or small values for the dependent variable, ordinary least squares (OLS) cannot be used to estimate the parameters β i . Instead, maximum likelihood techniques are used to maximize the value of the log likelihood (LL) function, which indicates how likely it is to obtain the observed values of Y, given the values of the independent variables and the parameters α, β1, ..., β k . Unlike OLS, which is able to solve directly for the parameters, the solution of the logistic regression model is found by iterating the estimation until the solution converges when the change in the likelihood function is negligible (for the present study, we used a threshold of 1×10−6, in the routine logitfit.m [53] for Matlab [49]).
Twice the negative of LL has approximately a χ2 distribution, which allows one to test the goodness of fit of a model. The value of −2LL for the logistic regression model with only the intercept included is designated D 0 to indicate that it is the −2 log likelihood statistic with none of the independent variables in the equation. It is analogous to the sum of squares (SST), in linear regression analysis. D m is analogous to the error sum of squares (SSE) in linear regression analysis, and is sometimes called "deviance", and is twice the negative LL function with the intercept as well as all the independent variables included. D m is used as an indicator of how poorly the model fits all of the independent variables in the equation. D m is analogous to the statistical significance of the unexplained variance in a regression model. The most direct analogue in logistic regression analysis to the regression sum of squares (SSR) in linear regression analysis is the difference between D 0 and D m:
G m is analogous to the multivariate F-test for linear regression, as well as the regression sum of squares. Treated as a χ2 statistic, G m provides a test of the null hypothesis that β1=β2=...=β k =0 for the logistic regression model. If G m is statistically significant (with, for example, p<0.05, a 95% confidence level), then the null hypothesis (of random correlation) is rejected and one can conclude that the model allows us to make predictions of P(Y=1).
A natural choice for comparing the strength of the relationship between variables is the analogy to R 2 as the sum of the squares of the residuals over the total sum of squares (SST), SST=SSR / SST, in a linear regression model. R L 2 is a proportional reduction in χ2 or a proportional reduction in the absolute value of the LL measure.
This statistic indicates by how much the inclusion of the independent variables in the model increases the goodness of fit D 0 to the χ2 statistic. R L 2 varies between 0 (for a model in which G m=0, D m=D 0 and the independent variables are useless in predicting the dependent variable) and 1 (for a model in which G m=−2LL and D m=0 and the model predicts the dependent variable with perfect accuracy).
Rights and permissions
About this article
Cite this article
García-Sosa, A.T., Mancera, R.L. & Dean, P.M. WaterScore: a novel method for distinguishing between bound and displaceable water molecules in the crystal structure of the binding site of protein-ligand complexes. J Mol Model 9, 172–182 (2003). https://doi.org/10.1007/s00894-003-0129-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-003-0129-x