Abstract
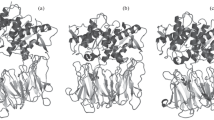
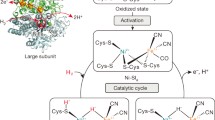
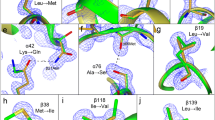
The aminopeptidase from Aeromonas proteolytica (AAP) contains two zinc ions in the active site and catalyzes the degradation of peptides. Herein we report the crystal structures of AAP at 0.95-Å resolution at neutral pH and at 1.24-Å resolution at low pH. The combination of these structures allowed the precise modeling of atomic positions, the identification of the metal bridging oxygen species, and insight into the physical properties of the metal ions. On the basis of these structures, a new putative catalytic mechanism is proposed for AAP that is likely relevant to all binuclear metalloproteases.





Similar content being viewed by others
Abbreviations
- AAP:
-
Aminopeptidase from Aeromonas proteolytica
- BuBA:
-
1-Butaneboronic acid
- CSD:
-
Cambridge Structural Database
- ESD:
-
Estimated standard deviation
- HEPES:
-
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- LPA:
-
l-Leucinephosphonic acid
- rms:
-
Root mean square
- Tris:
-
Tris(hydroxymethyl)aminomethane
References
Sträter N, Lipscomb WN, Klabunde T, Krebs B (1996) Angew Chem Int Ed Engl 35:2024
Wilcox D (1996) Binuclear metallohydrolases. Chem Rev 96:2435–2458
Lipscomb W, Sträter N (1996) Chem Rev 96:2375–2434
Dismukes G (1996) Chem Rev 96:2909–2926
Prescott J, Wilkes S (1976) Methods Enzymol 45:530–543
Stamper C, Bennett B, Edwards T, Holz R, Ringe D, Petsko G (2001) Biochemistry 40:7035–7046
Bzymek KP, Holz RC (2004) J Biol Chem 279:31018–31025
Desmarais WT, Bienvenue DL, Bzymek KP, Holz RC, Petsko GA, Ringe D (2002) Structure 8:1063–1072
Harding MM (1999) Acta Crystallogr D 55:1432–1443
Diaz N, Suarez D, Merz KM Jr (2000) J Am Chem Soc 122:4197–4208
Schalk C, Remy J, Chevrier B, Moras D, Tarnus C (1992) Arch Biochem Biophys 294:91–97
Chevrier B, Schalk C, D’Orchymont H, Rondeau J-M, Moras D, Tarnus C (1994) Structure 2:283–291
Otwinowski Z, Minor W (1997) Methods Enzymol 276:307–326
Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Acta Crystallogr D 54:905–921
Sheldrick G, Schneider T (1997) Methods Enzymol 277:319–343
Lamzin VS, Wilson KS (1993) Acta Crystallogr D 49:129–147
Holz RC (2002) Coord Chem Rev 232:5–26
Barrow GM (1981) Physical chemistry for the life sciences, 2nd edn. McGraw-Hill, New York, p 432
Christianson DW, Alexander SA (1989) J Am Chem Soc 111:6412–6419
Baker JO, Prescott JM (1983) Biochemistry 22:5322–5331
De Paola CC, Bennett B, Holz RC, Ringe D, Petsko GA (1999) Biochemistry 38:9048–9053
Acknowledgements
This work was supported by the National Science Foundation (CHE-0549221 to R.C.H.), by the National Institutes of Health (GM26788 to G.A.P. and D.R.), and, in part, by the Macromolecular Training Grant from the National Institutes of Health (W.D.).
Author information
Authors and Affiliations
Corresponding authors
Additional information
The coordinates for the 0.95-Å resolution structure and the 1.24-Å structure at pH 4.7 were deposited in the RCSB Protein Data Bank and have PDB ID numbers of 1RTQ and 2DEA, respectively.
Rights and permissions
About this article
Cite this article
Desmarais, W., Bienvenue, D.L., Bzymek, K.P. et al. The high-resolution structures of the neutral and the low pH crystals of aminopeptidase from Aeromonas proteolytica . J Biol Inorg Chem 11, 398–408 (2006). https://doi.org/10.1007/s00775-006-0093-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-006-0093-x