Abstract
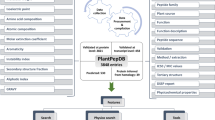
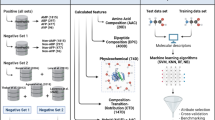
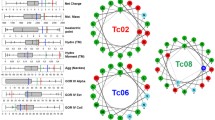
Emerging infectious diseases (EIDs) are a severe problem caused by fungi in human and plant species across the world. They pose a worldwide threat to food security as well as human health. Fungal infections are increasing now day by day worldwide, and the current antimycotic drugs are not effective due to the emergence of resistant strains. Therefore, it is an urgent need for the finding of new plant-origin antifungal peptides (PhytoAFPs). Huge numbers of peptides were extracted from different plant species which play a protective role against fungal infection. Hundreds of plant-origin peptides with antifungal activity have already been reported. So there is a requirement of a dedicated platform which systematically catalogs plant-origin peptides along with their antifungal properties. PlantAFP database is a resource of experimentally verified plant-origin antifungal peptides, collected from research articles, patents, and public databases. The current release of PlantAFP database contains 2585 peptide entries among which 510 are unique peptides. Each entry provides comprehensive information of a peptide that includes its peptide sequence, peptide name, peptide class, length of the peptide, molecular mass, antifungal activity, and origin of peptides. Besides this primary information, PlantAFP stores peptide sequences in SMILES format. In order to facilitate the user, many tools have been integrated into this database that includes BLAST search, peptide search, SMILES search, and peptide-mapping is also included in the database. PlantAFP database is accessible at http://bioinformatics.cimap.res.in/sharma/PlantAFP/.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/s0022-2836(05)80360-2
Barbosa Pelegrini P, del Sarto RP, Silva ON, Franco OL, Grossi-de-Sa MF (2011) Antibacterial peptides from plants: what they are and how they probably work. Biochem Res Int 2011
Bongomin F, Gago S, Oladele R, Denning D (2017) Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi 3(4):57
Brahmachary M, Krishnan S, Koh JLY, Khan AM, Seah SH, Tan TW, Brusic V, Bajic VB (2004) ANTIMIC: a database of antimicrobial sequences. Nucleic acids research 32(suppl_1):D586–D589
Broekaert WF, Terras F, Cammue B, Osborn RW (1995) Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol 108(4):1353
Craik DJ, Fairlie DP, Liras S, Price D (2013) The future of peptide-based drugs. Chem Biol Drug Design 81(1):136–147
De Beer A, Vivier MA (2011) Four plant defensins from an indigenous South African Brassicaceae species display divergent activities against two test pathogens despite high sequence similarity in the encoding genes. BMC Res Notes 4(1):459
De Lucca AJ, Walsh TJ (1999) Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrobial agents and chemotherapy 43(1):1–11
Devocelle M (2012) Targeted antimicrobial peptides. Front Immunol 3:309
Duncan VM, O’Neil DA (2013) Commercialization of antifungal peptides. Fungal Biol Rev 26(4):156–165
Fan L, Sun J, Zhou M, Zhou J, Lao X, Zheng H, Xu H (2016) DRAMP: a comprehensive data repository of antimicrobial peptides. Sci Rep 6:24482
Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484(7393):186
Fjell CD, Hancock RE, Cherkasov A (2007) AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics 23(9):1148–1155
Garvey M, Meehan S, Gras SL, Schirra HJ, Craik DJ, Van der Weerden NL, Anderson MA, Gerrard JA, Carver JA (2013) A radish seed antifungal peptide with a high amyloid fibril-forming propensity. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1834(8):1615–1623
Hammami R, Ben Hamida J, Vergoten G, Fliss I (2008) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic acids research 37(suppl_1):D963–D968
Keymanesh K, Soltani S, Sardari S (2009) Application of antimicrobial peptides in agriculture and food industry. World J Microbiol Biotechnol 25(6):933–944
Knogge W (1996) Fungal infection of plants. The Plant Cell 8(10):1711
Kuć J (1982) Induced immunity to plant disease. Bioscience 32(11):854–860
Lehrer RI, Ganz T (1999) Antimicrobial peptides in mammalian and insect host defence. Curr Opin Immunol 11(1):23–27
Loeza-Angeles H, Sagrero-Cisneros E, Lara-Zárate L, Villagómez-Gómez E, López-Meza JE, Ochoa-Zarzosa A (2008) Thionin Thi2. 1 from Arabidopsis thaliana expressed in endothelial cells shows antibacterial, antifungal and cytotoxic activity. Biotechnol Lett 30(10):1713
Marcos JF, Muñoz A, Pérez-Payá E, Misra S, López-García B (2008) Identification and rational design of novel antimicrobial peptides for plant protection. Annu Rev Phytopathol 46:273–301
Maróti G, Kereszt A, Kondorosi E, Mergaert P (2011) Natural roles of antimicrobial peptides in microbes, plants and animals. Res Microbiol 162(4):363–374
Meyer V, Andersen MR, Brakhage AA, Braus GH, Caddick MX, Cairns TC, Vries RP, Haarmann T, Hansen K, Hertz-Fowler C (2016) Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: a white paper. Fungal Biol Biotechnol 3(1):6
Montesinos E (2007) Antimicrobial peptides and plant disease control. FEMS Microbiol Lett 270(1):1–11
Neumann GM, Condron R, Polya GM (1996) Purification and mass spectrometry-based sequencing of yellow mustard (Sinapis alba L.) 6 kDa proteins Identification as antifungal proteins. Int J Peptide Protein Res 47(6):437–446
Omidvar R, Xia Y, Porcelli F, Bohlmann H, Veglia G (2016) NMR structure and conformational dynamics of AtPDFL2. 1, a defensin-like peptide from Arabidopsis thaliana. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1864(12):1739–1747
Osborn RW, De Samblanx GW, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees SB, Broekaert WF (1995) Isolation and characterisation of plant defensins from seeds of Asteraceae, Fabaceae Hippocastanaceae and Saxifragaceae. FEBS letters 368(2):257–262
Parfitt J, Barthel M, Macnaughton S (2010) Food waste within food supply chains: quantification and potential for change to 2050. Philos Trans R Soc B Biol Sci 365(1554):3065–3081
Rea MC, Ross RP, Cotter PD, Hill C (2011) Classification of bacteriocins from Gram-positive bacteria. Prokaryotic antimicrobial peptides. Springer, New York, pp 29–53
Remuzgo C, Oewel TS, Daffre S, Lopes TR, Dyszy FH, Schreier S, Machado-Santelli GM, Machini MT (2014) Chemical synthesis, structure–activity relationship, and properties of shepherin I: a fungicidal peptide enriched in glycine-glycine-histidine motifs. Amino Acids 46(11):2573–2586
Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46(11):941–950
Shi L, Zhang Q, Rui W, Lu M, Jing X, Shang T, Tang J (2004) BioPD: a web-based information center for bioactive peptides. Regul Peptides 120(1–3):1–3
Shuping D, Eloff JN (2017) The use of plants to protect plants and food against fungal pathogens: a review. Afr J Trad Complem Altern Med 14(4):120
Terras F, Schoofs H, De Bolle M, Van Leuven F, Rees SB, Vanderleyden J, Cammue B, Broekaert WF (1992a) Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. Journal of biological Chemistry 267 (22):15301-15309
Terras FR, Goderis IJ, Van Leuven F, Vanderleyden J, Cammue BP, Broekaert WF (1992b) In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid transfer proteins. Plant physiology 100 (2):1055-1058
Terras FR, Torrekens S, Van Leuven F, Osborn RW, Vanderleyden J, Cammue BP, Broekaert WF (1993) A new family of basic cysteine-rich plant antifungal proteins from Brassicaceae species. FEBS letters 316(3):233–240
Terras FR, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J (1995) Small cysteine-rich antifungal proteins from radish: their role in host defense. The Plant Cell 7(5):573–588
Thundimadathil J (2012) Cancer treatment using peptides: current therapies and future prospects. Journal of amino acids 2012
Tyagi A, Tuknait A, Anand P, Gupta S, Sharma M, Mathur D, Joshi A, Singh S, Gautam A, Raghava GP (2014) CancerPPD: a database of anticancer peptides and proteins. Nucleic acids research 43(D1):D837–D843
Vriens K, Cools TL, Harvey PJ, Craik DJ, Braem A, Vleugels J, De Coninck B, Cammue BP, Thevissen K (2016) The radish defensins RsAFP1 and RsAFP2 act synergistically with caspofungin against Candida albicans biofilms. Peptides 75:71–79
Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S (2014) CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic acids research 42(D1):D1154–D1158
Wang Z, Wang G (2004) APD: the antimicrobial peptide database. Nucleic Acids Res 32(suppl_1):D590–D592
Wang G, Li X, Wang Z (2008) APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res 37(suppl_1):D933–D937
Wang G, Li X, Wang Z (2015) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44(D1):D1087–D1093
Wang J, Yin T, Xiao X, He D, Xue Z, Jiang X, Wang Y (2018) StraPep: a structure database of bioactive peptides. Database 2018
Zareie R, Melanson DL, Murphy PJ (2002) Isolation of fungal cell wall degrading proteins from barley (Hordeum vulgare L.) leaves infected with Rhynchosporium secalis. Mol Plant Microbe Interact 15(10):1031–1039
Acknowledgements
The authors gratefully acknowledge the support provided by the CSIR- Central Institute of Medicinal and Aromatic Plants (CIMAP) and Jawaharlal Nehru University (JNU), New Delhi. Author (Atul Tyagi) is thankful to Indian Council of Medical Research (Project No. BIC/11(34)/2014) New Delhi, India, for ICMR-SRF fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
The article does not contain any studies in patients/animal by any of the authors.
Informed consent
The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript.
Additional information
Handling Editor: A. G. de Brevern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tyagi, A., Pankaj, V., Singh, S. et al. PlantAFP: a curated database of plant-origin antifungal peptides. Amino Acids 51, 1561–1568 (2019). https://doi.org/10.1007/s00726-019-02792-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-019-02792-5