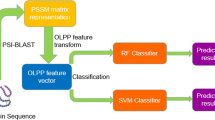
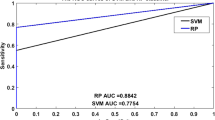
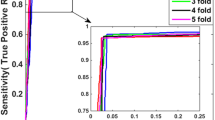
Abstract
Identifying protein–protein interactions (PPIs) is critical for understanding the cellular function of the proteins and the machinery of a proteome. Data of PPIs derived from high-throughput technologies are often incomplete and noisy. Therefore, it is important to develop computational methods and high-quality interaction dataset for predicting PPIs. A sequence-based method is proposed by combining correlation coefficient (CC) transformation and support vector machine (SVM). CC transformation not only adequately considers the neighboring effect of protein sequence but describes the level of CC between two protein sequences. A gold standard positives (interacting) dataset MIPS Core and a gold standard negatives (non-interacting) dataset GO-NEG of yeast Saccharomyces cerevisiae were mined to objectively evaluate the above method and attenuate the bias. The SVM model combined with CC transformation yielded the best performance with a high accuracy of 87.94% using gold standard positives and gold standard negatives datasets. The source code of MATLAB and the datasets are available on request under smgsmg@mail.ustc.edu.cn.


Similar content being viewed by others
References
Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW (2001) BIND-the biomolecular interaction network database. Nucleic Acids Res 29:242–245
Baudat G, Anouar F (2000) Generalized discriminant analysis using a kernel approach. Neural Comput 12:2385–2404
Ben-Hur A, Noble WS (2006) Choosing negative examples for the prediction of protein–protein interactions. BMC Bioinformatics 7:S2
Brenner SE, Chothia C, Hubbard TJ (1998) Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc Natl Acad Sci USA 95:6073–6078
Charton M, Charton BI (1982) The structural dependence of amino acid hydrophobicity parameters. J Theor Biol 99:629–644
Chothia C (1976) The nature of the accessible and buried surfaces in proteins. J Mol Biol 105:1–12
Deane CM, Salwinski L, Xenarios I, Eisenberg D (2002) Protein interactions: Two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics 1:349–356
Efron B, Hastie T, Johnstone I, Tibshirani R (2004) Least angle regression. Ann Stat 32:407–499
Eisenberg D, McLachlan AD (1986) Solvation energy in protein folding and binding. Nature 319:199–203
Fauchere JL (1988) Amino acid side chain parameters for correlation studies in biology and pharmacology. Int J Pept Protein Res 32:269–278
Faulon JL, Misra M, Martin S, Sale K, Sapra R (2008) Genome scale enzyme-metabolites and drug-target interaction predictions using the signature molecular descriptor. Bioinformatics 24:225–233
Feng ZP, Zhang CT (2000) Prediction of membrane protein types based on the hydrophobic index of amino acids. J Protein Chem 19:269–275
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29:1189–1232
Garel JP (1973) Coefficients de partage d’aminoacides, nucleobases, nucleosides et nucleotides dans un systeme solvant salin. J Chromatogr 78:381–391
Gavin AC et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147
Giot L et al (2003) A protein interaction map of Drosophila melanogaster. Science 302:1727–1736
Gomez SM, Noble WS, Rzhetsky A (2003) Learning to predict protein–protein interactions. Bioinformatics 19:1875–1881
Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185:862–864
Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V (2006) MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res 34:D436–D441
Guo X et al (2006) Assessing semantic similarity measures for the characterization of human regulatory pathways. Bioinformatics 22:967–973
Guo J, Wu XM, Zhang DY, Lin K (2008a) Genome-wide inference of protein interaction sites: lessons from the yeast high-quality negative protein–protein interaction dataset. Nucleic Acids Res 36:2002–2011
Guo YZ, Yu LZ, Wen ZN, Li ML (2008b) Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res 36:3025–3030
Ho Y et al (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183
Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA 78:3824–3828
Horne DS (1988) Prediction of protein helix content from an autocorrelation analysis of sequence hydrophobicities. Biopolymers 27:451–477
Hutchens JO (1970) Heat capacities, absolute entropies, and entropies of formation of amino acids and related compounds. In: Sober HA (ed) Handbook of biochemistry, 2nd edn. Chemical Rubber Co., Cleveland, pp B60–B61
Ito T et al (2000) Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA 97:1143–1147
Ito T et al (2001) A comprehensive two-hybrid analysis to explore the yeast protein ineractome. Proc Natl Acad Sci USA 98:4569–4574
Janin J (1979) Surface and inside volumes in globular proteins. Nature 277:491–492
Jansen R, Gerstein M (2004) Analyzing protein function on a genomic scale: the importance of gold-standard positives and negatives for network prediction. Curr Opin Microbiol 7:535–545
Koji T, William SN (2004) Learning kernels from biological networks by maximizing entropy. Bioinformatics 20:i326–i333
Krogan NJ et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643
Leslie C, Eskin E, Noble WS (2002) The spectrum kernel: a string kernel for SVM protein classification. In: Proceedings of the Pacific symposium on biocomputing, New Jersey. World Scientific, Singapore, pp 564–575
Li S et al (2004) A map of the interactome network of the metazoan c elegans. Science 303:540–543
Madaoui H, Guerois R (2008) Coevolution at protein complex interfaces can be detected by the complementarity trace with important impact for predictive docking. Proc Natl Acad Sci USA 105:7708–7713
Manly KF, Nettleton D, Hwang JT (2004) Genomics, prior probability, and statistical tests of multiple hypotheses. Genome Res 14:997–1001
Martin S, Roe D, Faulon JL (2005) Predicting protein–protein interactions using signature products. Bioinformatics 21:218–226
Mewes HW et al (2006) MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res 34:D169–D172
Prabhakaran M, Ponnuswamy PK (1982) Shape and surface features of globular proteins. Macromolecules 15:314–320
Rain JC et al (2001) The protein–protein interaction map of Helicobacter pylori. Nature 409:211–215
Resnik P (1999) Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J Artif Intell Res 11:95–130
Saito R et al (2003) Construction of reliable protein–protein interaction networks with a new interaction generality measure. Bioinformatics 19:756–763
Scholkopf B, Smola A, Muller KR (1998) Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput 10:1299–1319
Shen JW et al (2007) Predicting protein–protein interactions based only on sequences information. Proc Natl Acad Sci USA 104:4337–4341
Sokal RR, Thomson BA (2006) Population structure inferred by local spatial autocorrelation: an example from an Amerindian tribal population. Am J Phys Anthropol 129:121–131
Sprinzak E, Margalit H (2001) Correlated sequence-signatures as markers of protein–protein interaction. J Mol Biol 311:681–692
Sweet RM, Eisenberg D (1983) Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol 171:479–488
Uetz P et al (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403:623–627
Vapnik V (1998) Statistical learning theory. Wiley, New York
Von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large scale data sets of protein–protein interactions. Nature 417:399–403
Wang JZ, Du ZD, Payattakool R, Yu PS, Chen CF (2007) A new method to measure the semantic similarity of GO terms. Bioinformatics 23:1274–1281
Wiwatwattana N, Landau CM, Cope GJ, Harp GA, Kumar A (2007) Organelle DB: an updated resource of eukaryotic protein localization and function. Nucleic Acids Res 35:D810–D814
Wold S et al (1993) DNA and peptide sequences and chemical processes mutlivariately modelled by principal component analysis and partial least-squares projections to latent structures. Anal Chim Acta 277:239–253
Wu X, Zhu L, Guo J, Zhang DY, Lin K (2006) Prediction of yeast protein–protein interaction network: insights from the gene ontology and annotations. Nucleic Acids Res 34:2137–2150
Xenarios I et al (2002) Dip, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30:303–305
Yeang CH, Haussler D (2007) Detecting coevolution in and among protein domains. PLoS Comput Biol 3:e211
Zhu H et al (2001) Global analysis of protein activities using proteome chips. Science 293:2101–2105
Acknowledgments
This work was supported by the grants of the National Science Foundation of China, Nos. 60472111 and 30570368, the grant from the National Basic Research Program of China (973 Program), No. 2007CB311002, the grants from the National High Technology Research and Development Program of China (863 Program), Nos. 2007AA01Z167 and 2006AA02Z309, the grant of Oversea Outstanding Scholars Fund of CAS, No. 2005-1-18, HFUT, No. 070403F and the Knowledge Innovation Program of the Chinese Academy of Sciences (0823A16121).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Shi, MG., Xia, JF., Li, XL. et al. Predicting protein–protein interactions from sequence using correlation coefficient and high-quality interaction dataset. Amino Acids 38, 891–899 (2010). https://doi.org/10.1007/s00726-009-0295-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-009-0295-y