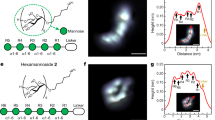
Abstract
As new methods to interrogate glycan organization on cells develop, it is important to have molecular level understanding of how chemical fixation can impact results and interpretations. Site-directed spin labeling technologies are well suited to study how the spin label mobility is impacted by local environmental conditions, such as those imposed by cross-linking effects of paraformaldehyde cell fixation methods. Here, we utilize three different azide-containing sugars for metabolic glycan engineering with HeLa cells to incorporate azido glycans that are modified with a DBCO-based nitroxide moiety via click reaction. Continuous wave X-band electron paramagnetic resonance spectroscopy is employed to characterize how the chronological sequence of chemical fixation and spin labeling impacts the local mobility and accessibility of the nitroxide-labeled glycans in the glycocalyx of HeLa cells. Results demonstrate that chemical fixation with paraformaldehyde can alter local glycan mobility and care should be taken in the analysis of data in any study where chemical fixation and cellular labeling occur.







Similar content being viewed by others
Availability of Data and Materials
ASCII files of all EPR and fluorescence data can be obtained by contacting the corresponding author directly.
References
A. Varki, Biological roles of glycans. Glycobiology 27, 3–49 (2017)
G. Wiederschain, Glycobiology and Human Diseases, Boca Taton: Taylor & Francis Group (2016)
X. Zhang, F.L. Kiechle, Glycosphingolipids in health and disease. Annu. Clin. Lab. Sci. 34, 3–13 (2004)
P.M. Rudd, Disease related glycosylation changes and biomarker discovery: challenges and possibilities in an emerging field (IOS Press, Fairfax, 2009)
R.L. Schnaar, A. Suzuki, P. Stanley, Glycosphingolipids, in Essentials of Glycobiology. ed. by A. Varki et al. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor p, 2008), pp.129–142
D. Russo, S. Parashuraman, G. D’Angelo, Glycosphingolipid–protein interaction in signal transduction. Int. J. Mol. Sci. 17(1–23), 1732 (2016)
C. Garcia-Ruiz, A. Morales, J.C. Fernandez-Checa, Glycosphingolipids and cell death: one aim, many ways. Apoptosis 20, 607–620 (2015)
P.H.H. Lopez, R.L. Schnaar, Gangliosides in cell recognition and membrane protein regulation. Curr. Opin. Struct. Biol. 19, 549–557 (2009)
C.-L. Schengrund, Gangliosides: glycosphingolipids essential for normal neural development and function. Trend. Biochem. Sci. 40, 397–406 (2015)
K. Palmano et al., The role of gangliosides in neurodevelopment. Nutrients 7, 3891–3913 (2015)
V. Gouaze-Andersson, M.C. Cabot, Glycosphingolipids and drug resistance. Biochim. Biophys. Acta 1758, 2096–2103 (2006)
S. Groux-Degroote, Y. Guerardel, P. Delannoy, Gangliosides: structures, biosynthesis, analysis, and roles in cancer. ChemBioChem 18, 1146–1154 (2017)
E.S. Qamsari et al., Ganglioside as a therapy target in various types of cancer. Asian Pac. J. Cancer Prev. 17, 1643–1647 (2016)
J. Inokuchi, GM3 and diabetes. Glycoconj. J. 31, 193–197 (2014)
R. Halmer, S. Walter, K. Faßbender, Sphingolipids: Important players in multiple sclerosis. Cell. Physiol. Biochem. 34, 111–118 (2014)
G. van Echten-Deckert, J. Walter, Sphingolipids: critical players in Alzheimer’s disease. Prog. Lipid Res. 51, 378–393 (2012)
T. Ariga, C. Wakade, R.K. Yu, The pathological roles of ganglioside metabolism in Alzheimer’s disease: Effects of gangliosides on neurogenesis. Int. J. Alzh. Dis. ID 193618: p. 1–14 (2011)
L. Ginzburg, Y. Kacher, K.H. Futerman, The pathogenesis of glycosphingolipid storage disorders. Semin. Cell Dev. Biol. 15, 417–431 (2004)
M.A.J. Ferguson, A.F. Williams, Cell-surface anchoring of proteins via glycosylphosphatidylinositol structure. Annu. Rev. Biochem. 57, 285–320 (1988)
P.J. Robinson, Signal transduction by GPI-anchored membrane proteins. Cell Biol. Intern. Rep. 15, 761–767 (1991)
H. Onda et al., A Novel secreted tumor antigen with a glycosylphosphatidylinositol-anchored structure ubiquitously expressed in human cancers. Biochem. Biophys. Res. Commun. 285(2), 235–243 (2001)
G. Wu et al., Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 66(20), 9829–9836 (2006)
P. Zhao et al., Proteomic identification of glycosylphosphatidylinositol anchor-dependent membrane proteins elevated in breast carcinoma. J. Biol. Chem. 287, 25230–25240 (2012)
S. Dolezala et al., Elevated levels of glycosylphosphatidylinositol (GPI) anchored proteins in plasma from human cancers detected by C. septicum alpha toxin. Cancer Biomark. 14, 55–62 (2014)
K. Sambamurti et al., Glycosylphosphatidylinositol-anchored proteins play an important role in the biogenesis of the Alzheimer’s amyloid beta-protein. J. Biol. Chem. 274(38), 26810–26814 (1999)
E.I. Walter et al., Effect of glycoinositolphospholipid anchor lipid groups on functional properties of decay-accelerating factor protein in cells. J. Biol. Chem. 267, 1245–1252 (1992)
M.A. McDowell, D.M. Ransom, J.D. Bangs, Glycosylphosphatidylinositol-dependent secretory transport in Trypanosoma brucei. Biochem. J. 335, 681–689 (1998)
P. Boccuni et al., Glycosyl phosphatidylinositol (GPI)-anchored molecules and the pathogenesis of paroxysmal nocturnal hemoglobinuria. Crit. Rev. Oncol. Hematol. 33, 25–43 (2000)
B. Wang, G.-J. Boons, Carbohydrate Recognition: Biological Problems, Methods, and Applications (Wiley, Hoboken, 2011)
M. Jaiswal et al., A metabolically engineered spin-labeling approach for studying glycans on cells. Chem. Sci. 11, 12522–12532 (2020)
M. Jaiswal et al., Enzymatic glycoengineering-based spin labelling of cell surface sialoglycans to enable their analysis by electron paramagnetic resonance (EPR) spectroscopy. Analyst 147(5), 784–788 (2022)
M. Jaiswal et al., Different biophysical properties of cell surface α2,3- and α2,6-sialoglycans revealed by electron paramagnetic resonance spectroscopic studies. J. Phys. Chem. B 127(8), 1749–1757 (2023)
W.L. Hubbell, D.S. Cafiso, C. Altenbach, Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7(9), 735–739 (2000)
W.L. Hubbell et al., Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 8(5), 649–656 (1998)
I.D. Sahu, G.A. Lorigan, Site-directed spin labeling EPR for studying membrane proteins. Biomed. Res. Int. 2018, 3248289 (2018)
P. Nguyen, P.Z. Qin, RNA dynamics: perspectives from spin labels. Wiley Interdiscip Rev RNA 3(1), 62–72 (2012)
P.Z. Qin et al., Monitoring RNA base structure and dynamics using site-directed spin labeling. Biochemistry 42(22), 6772–6783 (2003)
X. Zhang et al., Studying RNA using site-directed spin-labeling and continuous-wave electron paramagnetic resonance spectroscopy. Methods Enzymol. 469, 303–328 (2009)
D.S. Cafiso, Identifying and quantitating conformational exchange in membrane proteins using site-directed spin labeling. Acc. Chem. Res. 47(10), 3102–3109 (2014)
G.E. Fanucci, D.S. Cafiso, Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 16(5), 644–653 (2006)
L. Columbus, W.L. Hubbell, A new spin on protein dynamics. Trends Biochem. Sci. 27(6), 288–295 (2002)
L. Columbus, W.L. Hubbell, Mapping backbone dynamics in solution with site-directed spin labeling: GCN4-58 bZip free and bound to DNA. Biochemistry 43(23), 7273–7287 (2004)
L. Columbus et al., Molecular motion of spin labeled side chains in alpha-helices: analysis by variation of side chain structure. Biochemistry 40(13), 3828–3846 (2001)
Z. Zhang et al., Multifrequency electron spin resonance study of the dynamics of spin labeled T4 lysozyme. J. Phys. Chem. B 114(16), 5503–5521 (2010)
T.M. Casey et al., Continuous wave W- and D-Band EPR spectroscopy offer “sweet-spots” for characterizing conformational changes and dynamics in intrinsically disordered proteins. Biochem. Biophys. Res. Commun. 450(1), 723–728 (2014)
C. Altenbach et al., Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry 40(51), 15471–15482 (2001)
C.C. Jao et al., Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci U S A 105(50), 19666–19671 (2008)
M. Chen et al., Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 282(34), 24970–24979 (2007)
J. Pyka et al., Accessibility and dynamics of nitroxide side chains in T4 lysozyme measured by saturation recovery EPR. Biophys. J. 89(3), 2059–2068 (2005)
C. Altenbach et al., Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys. J. 89(3), 2103–2112 (2005)
C. Altenbach et al., A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA 91(5), 1667–1671 (1994)
T.L. Kirby, C.B. Karim, D.D. Thomas, Electron paramagnetic resonance reveals a large-scale conformational change in the cytoplasmic domain of phospholamban upon binding to the sarcoplasmic reticulum Ca-ATPase. Biochemistry 43(19), 5842–5852 (2004)
G.E. Fanucci et al., Structure and dynamics of the beta-barrel of the membrane transporter BtuB by site-directed spin labeling. Biochemistry 41(39), 11543–11551 (2002)
A. Gross, W.L. Hubbell, Identification of protein side chains near the membrane-aqueous interface: a site-directed spin labeling study of KcsA. Biochemistry 41(4), 1123–1128 (2002)
C.Y. Cheng et al., Hydration dynamics as an intrinsic ruler for refining protein structure at lipid membrane interfaces. Proc Natl Acad Sci USA 110(42), 16838–16843 (2013)
I. Kaminker, R. Barnes, S.I. Han, Overhauser dynamic nuclear polarization studies on local water dynamics. Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Pt B 564, 457–483 (2015)
C. Y. Cheng, et al. Cholesterol enhances surface water diffusion of phospholipid bilayers. J. Chem. Phys. 141(22), (2014)
J. Song, B. Allison, S. Han, Local water diffusivity as a molecular probe of surface hydrophilicity. MRS Bull. 39(12), 1082–1088 (2014)
J.M. Franck, J.A. Scott, S.I. Han, Nonlinear scaling of surface water diffusion with bulk water viscosity of crowded solutions. J. Am. Chem. Soc. 135(11), 4175–4178 (2013)
Y. Hong et al., Hydrophobicity of arginine leads to reentrant liquid-liquid phase separation behaviors of arginine-rich proteins. Nat. Commun. 13(1), 7326 (2022)
H. Moon et al., Evidence for entropically controlled interfacial hydration in mesoporous organosilicas. J. Am. Chem. Soc. 144(4), 1766–1777 (2022)
I.R. Smith et al., Probing membrane hydration at the interface of self-assembled peptide amphiphiles using electron paramagnetic resonance. ACS Macro Lett. 7(10), 1261–1266 (2018)
E. Saxon, C.R. Bertozzi, Cell surface engineering by a modified Staudinger reaction. Science 287(5460), 2007–2010 (2000)
R. Xie et al., In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc Natl Acad Sci USA 113(19), 5173–5178 (2016)
P.A. Amato, E.R. Unanue, D.L. Taylor, Distribution of actin in spreading macrophages: a comparative study on living and fixed cells. J. Cell Biol. 96(3), 750–761 (1983)
P. Lonn, U. Landegren, Close encounters - probing proximal proteins in live or fixed cells. Trends Biochem. Sci. 42(7), 504–515 (2017)
G.I. Malinin, T.I. Malinin, Effects of dimethylsulfoxide on the ultrastructure of fixed cells. Biotech. Histochem. 79(2), 65–69 (2004)
P. Watson, A.T. Jones, D.J. Stephens, Intracellular trafficking pathways and drug delivery: fluorescence imaging of living and fixed cells. Adv. Drug Deliv. Rev. 57(1), 43–61 (2005)
S.R. Yoshida, B.K. Maity, S. Chong, Visualizing protein localizations in fixed cells: caveats and the underlying mechanisms. J. Phys. Chem. B 127(19), 4165–4173 (2023)
S. Irgen-Gioro, et al. Fixation can change the appearance of phase separation in living cells. Elife, 11, (2022)
S. Kiyoto et al., Improved chemical fixation of lipid-secreting plant cells for transmission electron microscopy. Microscopy (Oxf) 71(4), 206–213 (2022)
C.A. Edechi et al., Comparison of fixation methods for the detection of claudin 1 and E-cadherin in breast cancer cell lines by immunofluorescence. J. Histochem. Cytochem. 70(2), 181–187 (2022)
T. Ichikawa et al., Chemical fixation creates nanoscale clusters on the cell surface by aggregating membrane proteins. Commun Biol 5(1), 487 (2022)
B. Cheng et al., 9-azido analogues of three sialic acid forms for metabolic remodeling of cell-surface sialoglycans. ACS Chem. Biol. 14(10), 2141–2147 (2019)
K. Ikeda et al., Chemoenzymatic synthesis of an N-acetylneuraminic acid analogue having a carbamoylmethyl group at C-4 as an inhibitor of sialidase from influenza virus. Carbohydr. Res. 312(4), 183–189 (1998)
C Wuebben, et al. Site-Directed Spin Labeling of RNA with a Gem-Diethylisoindoline Spin Label: PELDOR, Relaxation, and Reduction Stability. Molecules. 24(24), (2019)
S. Stoll, A. Schweiger, EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178(1), 42–55 (2006)
A.P. Todd, G.L. Millhauser, Esr-spectra reflect local and global mobility in a short spin-labeled peptide throughout the alpha-helix-]coil transition. Biochemistry 30(22), 5515–5523 (1991)
S.M. Miick, A.P. Todd, G.L. Millhauser, Position-dependent local motions in spin-labeled analogs of a short alpha-helical peptide determined by electron-spin-resonance. Biochemistry 30(39), 9498–9503 (1991)
N.L. Pirman et al., Characterization of the disordered-to-α-helical transition of IA3 by SDSL-EPR spectroscopy. Protein Sci. 20, 150–159 (2011)
S.J. Moons et al., Sialic acid glycoengineering using N-acetylmannosamine and sialic acid analogs. Glycobiology 29(6), 433–445 (2019)
P.Z. Qin, J. Iseri, A. Oki, A model system for investigating lineshape/structure correlations in RNA site-directed spin labeling. Biochem. Biophys. Res. Commun. 343(1), 117–124 (2006)
E.A. Hoffman et al., Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 290(44), 26404–26411 (2015)
E. Raczuk, et al., Different Schiff Bases-Structure, Importance and Classification. Molecules, 27(3), (2022)
Funding
This study was supported by NIGMS/NIH (R35GM131686; ZG), NSF (MCB-1715384; GEF), an NIH instrumentation grant (S10 RR031603) for the Bruker E500 spectrometer, and Department of Chemistry, University of Florida for the Magnettech MS-5000 benchtop spectrometer. ZG thanks Steven and Rebecca Scott endowment to support our research.
Author information
Authors and Affiliations
Contributions
MJ, JG, and SK contributed to molecule synthesis, cell culturing, glycoengineering, spin labeling and chemical fixation; TTT and MZ contributed to EPR studies and spectral fitting analyses; GEF and ZG were overall responsible for the project design and supervision. The manuscript was written through contributions of all authors, and all authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jaiswal, M., Tran, T.T., Guo, J. et al. Spin-Labeling Insights into How Chemical Fixation Impacts Glycan Organization on Cells. Appl Magn Reson 55, 317–333 (2024). https://doi.org/10.1007/s00723-023-01624-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-023-01624-w