Summary.
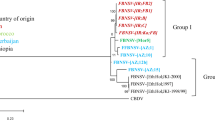
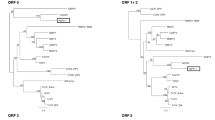
The complete nucleotide (nt) sequence of a Singapore isolate of broad bean wilt fabavirus from Megakepasma erythrochlamys L., designated BBWV-ME, was determined. Its bipartite genome consisted of two positive-sense single-stranded ribonucleic acids (RNA). RNA1 (5951 nt in length) encoded a putative protease cofactor, nucleotide triphosphate (NTP)-binding domain (helicase), viral genome-linked protein (VPg), protease and RNA-dependent RNA polymerase (RdRp). RNA2 (3607 nt in length) encoded a putative movement protein (MP) and coat proteins (CP). Genome organization of BBWV-ME was similar to other viruses in the Comoviridae family. Phylogenetic analyses showed that fabaviruses were more closely related to the comoviruses than the nepoviruses.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received February 23, 2000 Accepted July 18, 2000
Rights and permissions
About this article
Cite this article
Koh, L., Cooper, J. & Wong, S. Complete sequences and phylogenetic analyses of a Singapore isolate of broad bean wilt fabavirus. Arch. Virol. 146, 135–147 (2001). https://doi.org/10.1007/s007050170197
Issue Date:
DOI: https://doi.org/10.1007/s007050170197