Abstract
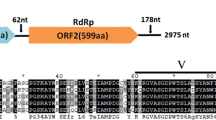
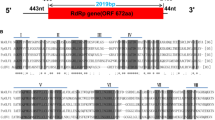
A novel polymycovirus isolated from the plant-pathogenic fungus Colletotrichum gloeosporioides was identified. The viral genome is composed of nine double-stranded RNA segments, ranging in size from 699 bp to 2,444 bp. With the exception of dsRNA5, which contains two open reading frames (ORF5-1 and ORF5-2), the other dsRNA segments each contain one ORF. The proteins encoded by ORFs 1–8 are homologous to the proteins encoded by ORFs 1–8 of Colletotrichum camelliae filamentous virus 1 (CcFV-1). The amino acid sequences of the RNA-dependent RNA polymerase (RdRp) encoded by ORF1 and the viral methyltransferase encoded by ORF3 share 87.6% and 83.3% identity with CcFV-1. The proline-alanine-serine-rich protein (PASrp) encoded by ORF4 shares 86.6% sequence identity with that of CcFV-1. The proteins encoded by ORFs 2, 5 − 1, 6, 7, and 8 share 86.6%, 82.5%, 89.0%, 45.7%, and 95.5% sequence identity, respectively, with the corresponding proteins of CcFV-1. dsRNA9 is a defective copy of dsRNA2 that lacks a stretch of 1556 bp (nt 519 to nt 2074). Phylogenetic analysis based on the RdRp protein indicated that the novel virus clustered with members of the family Polymycoviridae, and based on the above results, we have tentatively named it "Colletotrichum gloeosporioides polymycovirus virus 1" (CgPmV1). To our knowledge, this is the first report of a polymycovirus with a defective dsRNA genome in C. gloeosporioides.



Similar content being viewed by others
References
Hollings M (1962) Viruses associated with A die-back disease of cultivated mushroom. Nature 196:962–965. https://doi.org/10.1038/196962a0
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368. https://doi.org/10.1016/j.virol.2015.02.034
Sato Y, Jamal A, Kondo H, Suzuki N (2020) Molecular characterization of a novel polymycovirus from Penicillium janthinellum with a focus on its genome-associated PASrp. Front Microbiol 11:592789. https://doi.org/10.3389/fmicb.2020.592789
Kang Q, Li L, Li J, Zhang S, Xie J, Li Q, Zhang Z (2021) A novel polymycovirus with defective RNA isolated from the entomopathogenic fungus Beauveria bassiana Vuillemin. Arch Virol 166:3487–3492. https://doi.org/10.1007/s00705-021-05238-0
Ma G, Wu C, Li Y, Mi Y, Zhou T, Zhao C, Wu X (2022) Identification and genomic characterization of a novel polymycovirus from Alternaria alternata causing watermelon leaf blight. Arch Virol 167:223–227. https://doi.org/10.1007/s00705-021-05272-y
Nerva L, Forgia M, Ciuffo M, Chitarra W, Chiapello M, Vallino M, Varese GC, Turina M (2019) The mycovirome of a fungal collection from the sea cucumber Holothuria polii. Virus Res 273:197737. https://doi.org/10.1016/j.virusres.2019.197737
Mahillon M, Decroës A, Liénard C, Bragard C, Legrève A (2019) Full genome sequence of a new polymycovirus infecting Fusarium redolens. Arch Virol 164:2215–2219. https://doi.org/10.1007/s00705-019-04301-1
Niu Y, Yuan Y, Mao J, Yang Z, Cao Q, Zhang T, Wang S, Liu D (2018) Characterization of two novel mycoviruses from Penicillium digitatum and the related fungicide resistance analysis. Sci Rep 8:5513. https://doi.org/10.1038/s41598-018-23807-3
Kotta-Loizou I, Coutts RH (2017) Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog 13:e1006183. https://doi.org/10.1371/journal.ppat.1006183
Jia H, Dong K, Zhou L, Wang G, Hong N, Jiang D, Xu W (2017) A dsRNA virus with filamentous viral particles. Nat Commun 8:168. https://doi.org/10.1038/s41467-017-00237-9
Cannon PF, Damm U, Johnston PR, Weir BS (2012) Colletotrichum - current status and future directions. Stud Mycol 73:181–213. https://doi.org/10.3114/sim0014
Agostini JP, Timmer LW, Mitchell DJ (1992) Morphological and pathological characteristics of strains of Colletotrichum gloeosporioides from citrus. Phytopathology 82:1377–1382. https://doi.org/10.1094/Phyto-82-1377
Zhong J, Pang XD, Zhu HJ, Gao BD, Huang WK, Zhou Q (2016) Molecular characterization of a trisegmented mycovirus from the plant pathogenic fungus Colletotrichum gloeosporioides. Viruses 8:268. https://doi.org/10.3390/v8100268
Wang Y, Liu S, Zhu HJ, Zhong J (2019) Molecular characterization of a novel mycovirus from the plant pathogenic fungus Colletotrichum gloeosporioides. Arch Virol 164:2859–2863. https://doi.org/10.1007/s00705-019-04354-2
Guo J, Zhu JZ, Zhou XY, Zhong J, Li CH, Zhang ZG, Zhu HJ (2019) A novel ourmia-like mycovirus isolated from the plant pathogenic fungus Colletotrichum gloeosporioides. Arch Virol 164:2631–2635. https://doi.org/10.1007/s00705-019-04346-2
Zhai L, Zhang M, Hong N, Xiao F, Fu M, Xiang J, Wang G (2018) Identification and characterization of a novel hepta-segmented dsRNA virus from the phytopathogenic fungus Colletotrichum fructicola. Front Microbiol 9:754. https://doi.org/10.3389/fmicb.2018.00754
Zhong J, Chen D, Lei XH, Zhu HJ, Zhu JZ, Da Gao B (2014) Detection and characterization of a novel gammapartitivirus in the phytopathogenic fungus Colletotrichum acutatum strain HNZJ001. Virus Res 190:104–109. https://doi.org/10.1016/j.virusres.2014.05.028
Rosseto P, Costa AT, Polonio JC, Da Silva AA, Pamphile JA, Azevedo JL (2016) Investigation of mycoviruses in endophytic and phytopathogenic strains of Colletotrichum from different hosts. Genet Mol Res 15:15017651. https://doi.org/10.4238/gmr.15017651
Marzano SL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL (2016) Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863. https://doi.org/10.1128/jvi.00357-16
Morris TJ, Dodds JA (1979) Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858. https://doi.org/10.1094/phyto-69-854
Li CX, Zhu JZ, Gao BD, Zhu HJ, Zhou Q, Zhong J (2019) Characterization of a novel ourmia-like mycovirus infecting Magnaporthe oryzae and implications for viral diversity and evolution. Viruses 11:223. https://doi.org/10.3390/v11030223
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Marchler-Bauer A, Bo Y, Han L et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Sato Y, Shamsi W, Jamal A, Bhatti MF, Kondo H, Suzuki N (2020) Hadaka virus 1: a capsidless eleven-segmented positive-sense single-stranded RNA virus from a phytopathogenic fungus. Fusarium oxysporum mBio 11:e00450–e00420. https://doi.org/10.1128/mBio.00450-20
Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N (2013) Effects of defective interfering RNA on symptom induction by, and replication of, a novel partitivirus from a phytopathogenic fungus, Rosellinia necatrix. J Virol 87:2330–2341. https://doi.org/10.1128/jvi.02835-12
Funding
This research was supported by the Hunan Provincial Natural Science Foundation of China (2021JJ30356), Hunan Provincial Key Research and Development Program of China (2020NK2045), and Hunan Provincial Science and Technology Innovation Platform Construction Fund (20K071).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor Ioly Kotta-Loizou
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, J., Xie, F., Zhang, Z. et al. Molecular characterization of a novel polymycovirus identified in the phytopathogenic fungus Colletotrichum gloeosporioides. Arch Virol 167, 2805–2810 (2022). https://doi.org/10.1007/s00705-022-05591-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05591-8