Abstract
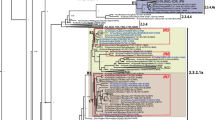
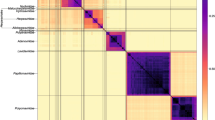
African swine fever is one of the most feared infectious diseases in the pig industry. African swine fever virus (ASFV) is an enveloped, cytoplasmic double-stranded DNA virus and the only member of the family Asfarviridae. Although ASFV is known to have been circulating on the African continent since at least 1921, little is known about the genetic characteristics of historical ASFV strains isolated in sub-Saharan Africa. The few complete ASFV genome sequences obtained from African historical isolates have demonstrated genetic diversity, but the available data are limited and insufficient for fully understanding the molecular evolution and continental spread of ASFV. Here, we report the complete genome sequence of the virulent ASFV strain K49, collected during an outbreak in the Belgian Congo (now the Democratic Republic of the Congo) in 1949. The complete genome sequence of ASFV strain K49 was determined using an Illumina HiSeq platform and is 189,523 bp in length with a mean GC content of 38.43%, with 189 genes annotated. This is the first reported complete genome sequence of an ASFV serogroup 2 isolate. Phylogenetic analysis demonstrated genetic divergence within genotype I, and strain K49 formed a separate branch from other ASFV genotype I isolates.


Similar content being viewed by others
References
Bisimwa PN, Ongus JR, Tiambo CK et al (2020) First detection of African swine fever (ASF) virus genotype X and serogroup 7 in symptomatic pigs in the Democratic Republic of Congo. Virol J 17:135. https://doi.org/10.1186/s12985-020-01398-8
Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17(11):1519–1533. https://doi.org/10.1089/cmb.2009.0238
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641. https://doi.org/10.1093/nar/27.23.4636
Dixon LK, Escribano JM, Martins C, Rock DL, Salas ML, Wilkinson PJ (2005) In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U (eds) Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press, London, UK, pp 135–143
Eustace Montgomery R (1921) On a form of swine fever occurring in British East Africa (Kenya Colony). J Comp Pathol Ther 34:159–191. https://doi.org/10.1016/s0368-1742(21)80031-4
Malogolovkin A, Burmakina G, Titov I et al (2015) Comparative analysis of African swine fever virus genotypes and serogroups. Emerg Infect Dis 21(2):312–315. https://doi.org/10.3201/eid2102.140649
Malogolovkin A, Burmakina G, Tulman ER, Delhon G, Diel DG et al (2015) African swine fever virus CD2v and C-type lectin gene loci mediate serological specificity. J Gen Virol 96:866–873. https://doi.org/10.1099/jgv.0.000024
Mulumba-Mfumu LK, Achenbach JE, Mauldin MR et al (2017) Genetic assessment of African swine fever isolates involved in outbreaks in the Democratic Republic of Congo between 2005 and 2012 reveals co-circulation of p72 genotypes I, IX and XIV, including 19 variants. Viruses 9(2):31. https://doi.org/10.3390/v9020031
Quembo CJ, Jori F, Vosloo W, Heath L (2017) Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound Emerg Dis 65:420–431. https://doi.org/10.1111/tbed.12700
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16(6):276–277. https://doi.org/10.1016/s0168-9525(00)02024-2
Titov I, Burmakina G, Morgunov Y, Morgunov S, Koltsov A, Malogolovkin A, Kolbasov D (2017) Virulent strain of African swine fever virus eclipses its attenuated derivative after challenge. Arch Virol 162(10):3081–3088. https://doi.org/10.1007/s00705-017-3471-5
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10(3):512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547. https://doi.org/10.1093/molbev/msy096
Acknowledgements
We are grateful to the staff of the Federal Research Center for Virology and Microbiology for the isolation and characterization of ASFV strain K49.
Funding
This research was funded by the Russian Science Foundation (Grant no. 20-76-10030).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Koltsov, A., Tulman, E.R., Namsrayn, S. et al. Complete genome sequence of virulent genotype I African swine fever virus strain K49 from the Democratic Republic of the Congo, isolated from a domestic pig (Sus scrofa domesticus). Arch Virol 167, 2377–2380 (2022). https://doi.org/10.1007/s00705-022-05543-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05543-2