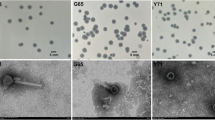
Abstract
Aeromonas salmonicida strains cause problematic bacterial infections in the aquaculture industry worldwide. The genus Aeromonas includes both mesophilic and psychrophilic species. Bacteriophages that infect Aeromonas spp. strains are usually specific for mesophilic or psychrophilic species; only a few bacteriophages can infect both types of strains. In this study, we characterized the podophage T7-Ah, which was initially found to infect the Aeromonas salmonicida HER1209 strain. The burst size of T7-Ah against its original host is 72 new virions per infected cell, and its burst time is 30 minutes. It has been found that this phage can lyse both mesophilic and psychrophilic A. salmonicida strains, as well as one strain of Escherichia coli. Its genome comprises 40,153 bp of DNA and does not contain any recognizable toxin or antibiotic resistance genes. The adsorption rate of the phage on highly sensitive bacterial strains was variable and could not be related to the presence or absence of a functional A-layer on the surface of the bacterial strains. The lipopolysaccharide migration patterns of both resistant and sensitive bacterial strains were also studied and compared to investigate the nature of the potential receptor of this phage on the bacterial surface. This study sheds light on the surprising diversity of lifestyles of the bacterial strains sensitive to phage T7-Ah and opens the door to the potential use of this phage against A. salmonicida infections in aquaculture.





Similar content being viewed by others
References
Austin B, Austin DA (2016) Bacterial fish pathogens—disease of farmed and wild fish, 6th edn. Springer International Publishing, Cham
Merino S, Rubires X, Knøchel S, Tomás JM (1995) Emerging pathogens: Aeromonas spp. Int J Food Microbiol 28:157–168. https://doi.org/10.1016/0168-1605(95)00054-2
Schuetz AN (2019) Emerging agents of gastroenteritis: Aeromonas, Plesiomonas, and the diarrheagenic pathotypes of Escherichia coli. Semin Diagn Pathol 36:187–192. https://doi.org/10.1053/j.semdp.2019.04.012
Vincent AT, Fernández-Bravo A, Sanchis M et al (2019) Investigation of the virulence and genomics of Aeromonas salmonicida strains isolated from human patients. Infect Genet Evol 68:1–9. https://doi.org/10.1016/j.meegid.2018.11.019
Vincent AT, Trudel MV, Freschi L et al (2016) Increasing genomic diversity and evidence of constrained lifestyle evolution due to insertion sequences in Aeromonas salmonicida. BMC Genomics 17:44. https://doi.org/10.1186/s12864-016-2381-3
Pavan ME, Abbott SL, Zorzo J, Janda JM (2000) Aeromonas salmonicida subsp. pectinolytica subsp. nov., a new pectinase-positive subspecies isolated from a heavily polluted river. Int J Syst Evol Microbiol 50:1119–1124. https://doi.org/10.1099/00207713-50-3-1119
Vincent AT, Rouleau FD, Moineau S, Charette SJ (2017) Study of mesophilic Aeromonas salmonicida A527 strain sheds light on the species’ lifestyles and taxonomic dilemma. FEMS Microbiol Lett 364:fnx239. https://doi.org/10.1093/femsle/fnx239
Vincent AT, Bernatchez A, Frey J, Charette SJ (2019) A mesophilic Aeromonas salmonicida strain isolated from an unsuspected host, the migratory bird pied avocet. Microorganisms 7:592. https://doi.org/10.3390/microorganisms7120592
Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ (2016) Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis 16:e127–e133. https://doi.org/10.1016/S1473-3099(16)00100-6
Vincent AT, Paquet VE, Moineau S, Charette SJ (2019) The rise and fall of antibiotics in aquaculture. In: Derome N (ed) Microbial communities in aquaculture ecosystems. Springer, Cham, pp 8–26
Trudel MV, Vincent AT, Attéré SA et al (2016) Diversity of antibiotic-resistance genes in Canadian isolates of Aeromonas salmonicida subsp. salmonicida: dominance of pSN254b and discovery of pAsa8. Sci Rep 6:35617. https://doi.org/10.1038/srep35617
Culot A, Grosset N, Gautier M (2019) Overcoming the challenges of phage therapy for industrial aquaculture: a review. Aquaculture 513:734423. https://doi.org/10.1016/j.aquaculture.2019.734423
Schulz P, Pajdak-Czaus J, Robak S et al (2019) Bacteriophage-based cocktail modulates selected immunological parameters and post-challenge survival of rainbow trout (Oncorhynchus mykiss). J Fish Dis 42:1151–1160. https://doi.org/10.1111/jfd.13026
Le TS, Nguyen TH, Vo HP et al (2018) Protective effects of bacteriophages against Aeromonas hydrophila species causing motile Aeromonas septicemia (MAS) in striped catfish. Antibiotics 7:16. https://doi.org/10.3390/antibiotics7010016
Silva YJ, Moreirinha C, Pereira C et al (2016) Biological control of Aeromonas salmonicida infection in juvenile Senegalese sole (Solea senegalensis) with Phage AS-A. Aquaculture 450:225–233. https://doi.org/10.1016/j.aquaculture.2015.07.025
Kotob MH, Menanteau-Ledouble S, Kumar G et al (2016) The impact of co-infections on fish: a review. Vet Res 47:98. https://doi.org/10.1186/s13567-016-0383-4
Chandrarathna HPSU, Nikapitiya C, Dananjaya SHS et al (2018) Outcome of co-infection with opportunistic and multidrug resistant Aeromonas hydrophila and A. veronii in zebrafish: identification, characterization, pathogenicity and immune responses. Fish Shellfish Immunol 80:573–581. https://doi.org/10.1016/j.fsi.2018.06.049
Duman M, Saticioglu IB, Janda JM, Altun S (2018) The determination of the infectious status and prevalence of motile Aeromonas species isolated from disease cases in rainbow trout (Oncorhynchus mykiss) and aquarium fish. J Fish Dis 41:1843–1857. https://doi.org/10.1111/jfd.12896
Chow MS, Rouf MA (1983) Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl Environ Microbiol 45:1670–1676. https://doi.org/10.1128/aem.45.5.1670-1676.1983
Bai M, Cheng YH, Sun XQ et al (2019) Nine novel phages from a plateau lake in southwest China: insights into Aeromonas phage diversity. Viruses 11:615. https://doi.org/10.3390/v11070615
Kim JH, Son JS, Choresca CH et al (2012) Complete genome sequence of bacteriophage phiAS7, a T7-Like virus that infects Aeromonas salmonicida subsp. salmonicida. J Virol 86:2894–2895. https://doi.org/10.1128/jvi.07131-11
Anand T, Bera BC, Virmani N et al (2018) Isolation and characterization of a novel, T7-like phage against Aeromonas veronii. Virus Genes 54:160–164. https://doi.org/10.1007/s11262-017-1517-0
Bin WJ, Lin NT, Tseng YH, Weng SF (2016) Genomic characterization of the novel Aeromonas hydrophila phage Ahp1 suggests the derivation of a new subgroup from phiKMV-Like family. PLoS ONE 11:e0162060. https://doi.org/10.1371/journal.pone.0162060
Islam MS, Raz A, Liu Y et al (2019) Complete genome sequence of Aeromonas phage ZPAH7 with halo zones, isolated in China. Microbiol Resour Announc 8:e01678-e1718. https://doi.org/10.1128/MRA.01678-18
Cao Y, Li S, Wang D et al (2019) Genomic characterization of a novel virulent phage infecting the Aeromonas hydrophila isolated from rainbow trout (Oncorhynchus mykiss). Virus Res 273:197764. https://doi.org/10.1016/j.virusres.2019.197764
Paquet VE, Vincent AT, Moineau S, Charette SJ (2019) Beyond the A-layer: adsorption of lipopolysaccharides and characterization of bacteriophage-insensitive mutants of Aeromonas salmonicida subsp. salmonicida. Mol Microbiol 112:667–677. https://doi.org/10.1111/mmi.14308
Dallaire-Dufresne S, Tanaka KH, Trudel MV et al (2014) Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet Microbiol 169:1–7. https://doi.org/10.1016/j.vetmic.2013.06.025
Ishiguro EE, Ainsworth T, Harkness RE et al (1984) A temperate bacteriophage specific for strains of Aeromonas salmonicida possessing A-layer, a cell surface virulence factor. Curr Microbiol 10:199–202. https://doi.org/10.1007/BF01627255
Merino S, Camprubi S, Tomas JM (1992) Characterization of an O-antigen bacteriophage from Aeromonas hydrophila. Can J Microbiol 38:235–240. https://doi.org/10.1139/m92-040
Merino S, Camprubi S, Tomás JM (1990) Isolation and characterization of bacteriophage PM3 from Aeromonas hydrophila the bacterial receptor for which is the monopolar flagellum. FEMS Microbiol Lett 69:277–282. https://doi.org/10.1016/0378-1097(90)90080-a
Vincent AT, Paquet VE, Bernatchez A et al (2017) Characterization and diversity of phages infecting Aeromonas salmonicida subsp. salmonicida. Sci Rep 7:7054. https://doi.org/10.1038/s41598-017-07401-7
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. https://doi.org/10.1007/978-1-84882-087-6_9
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277. https://doi.org/10.1016/S0168-9525(00)02024-2
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr arXiv13033997
Li H, Handsaker B, Wysoker A et al (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Walker BJ, Abeel T, Shea T et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. https://doi.org/10.1371/journal.pone.0112963
Wattam AR, Davis JJ, Assaf R et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource. Nucleic Acids Res 45:D535–D542. https://doi.org/10.1093/nar/gkw1017
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Overbeek R, Olson R, Pusch GD et al (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. https://doi.org/10.1093/nar/gkt1226
Daher RK, Filion G, Tan SGE et al (2011) Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet Microbiol 152:353–360. https://doi.org/10.1016/j.vetmic.2011.04.034
Charette SJ, Brochu F, Boyle B et al (2012) Draft genome sequence of the virulent strain 01–B526 of the fish pathogen Aeromonas salmonicida. J Bacteriol 194:722–723. https://doi.org/10.1128/JB.06276-11
Dunn JJ, Studier FW, Gottesman M (1983) Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol 166:477–535. https://doi.org/10.1016/S0022-2836(83)80282-4
Liu J, Gao S, Dong Y et al (2020) Isolation and characterization of bacteriophages against virulent Aeromonas hydrophila. BMC Microbiol 20:141. https://doi.org/10.1186/s12866-020-01811-w
Groman NB, Suzuki G (1962) Temperature and lambda phage reproduction. J Bacteriol 84:431–437. https://doi.org/10.1128/JB.84.3.431-437.1962
Shan J, Korbsrisate S, Withatanung P et al (2014) Temperature dependent bacteriophages of a tropical bacterial pathogen. Front Microbiol 5:599. https://doi.org/10.3389/fmicb.2014.00599
Sanders ME, Klaenhammer TR (1984) Phage resistance in a phage-insensitive strain of Streptococcus lactis: temperature-dependent phage development and host-controlled phage replication. Appl Environ Microbiol 47:979–985. https://doi.org/10.1128/aem.47.5.979-985.1984
O’Driscoll J, Glynn F, Cahalane O et al (2004) Lactococcal plasmid pNP40 encodes a novel, temperature-sensitive restriction-modification system. Appl Environ Microbiol 70:5546–5556. https://doi.org/10.1128/AEM.70.9.5546-5556.2004
Kim JH, Son JS, Choi YJ et al (2012) Isolation and characterization of a lytic Myoviridae bacteriophage PAS-1 with broad infectivity in Aeromonas salmonicida. Curr Microbiol 64:418–426. https://doi.org/10.1007/s00284-012-0091-x
Kim JH, Son JS, Choi YJ et al (2012) Complete genome sequence and characterization of a broad-host range T4-like bacteriophage phiAS5 infecting Aeromonas salmonicida subsp. salmonicida. Vet Microbiol 157:164–171. https://doi.org/10.1016/j.vetmic.2011.12.016
Seed KD, Bodi KL, Kropinski AM et al (2011) Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. MBio 2:e00334-e410. https://doi.org/10.1128/mBio.00334-10.Editor
Mateus L, Costa L, Silva YJ et al (2014) Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 424–425:167–173. https://doi.org/10.1016/j.aquaculture.2014.01.001
Xia X, Yuen KY (2005) Differential selection and mutation between dsDNA and ssDNA phages shape the evolution of their genomic AT percentage. BMC Genet 6:20. https://doi.org/10.1186/1471-2156-6-20
Prabhakaran R, Chithambaram S, Xia X (2014) Aeromonas phages encode tRNAs for their overused codons. Int J Comput Biol Drug Des 7:168–182. https://doi.org/10.1504/IJCBDD.2014.061645
Gulla S, Lund V, Kristoffersen AB et al (2016) vapA (A-layer) typing differentiates Aeromonas salmonicida subspecies and identifies a number of previously undescribed subtypes. J Fish Dis 39:329–342. https://doi.org/10.1111/jfd.12367
Merino S, De Mendoza E, Canals R, Tomás JM (2015) Functional genomics of the Aeromonas salmonicida lipopolysaccharide O-antigen and A-layer from typical and atypical strains. Mar Drugs 13:3791–3808. https://doi.org/10.3390/md13063791
Austin B, Austin DA, Dalsgaard I et al (1998) Characterization of atypical Aeromonas salmonicida by different methods. Syst Appl Microbiol 21:50–64. https://doi.org/10.1016/S0723-2020(98)80008-8
Lund V, Espelid S, Mikkelsen H (2003) Vaccine efficacy in spotted wolffish Anarhichas minor: relationship to molecular variation in A-layer protein of atypical Aeromonas salmonicida. Dis Aquat Organ 56:31–42. https://doi.org/10.3354/dao056031
Kostrzynska M, Dooley JSG, Shimojo T et al (1992) Antigenic diversity of the S-layer proteins from pathogenic strains of Aeromonas hydrophila and Aeromonas veronii biotype sobria. J Bacteriol 174:40–47. https://doi.org/10.1128/jb.174.1.40-47.1992
Murray RGE, Dooley JSG, Whippey PW, Trust TJ (1988) Structure of an S layer on a pathogenic strain of Aeromonas hydrophila. J Bacteriol 170:2625–2630. https://doi.org/10.1128/jb.170.6.2625-2630.1988
Chu S, Cavaignac S, Feutrier J et al (1991) Structure of the tetragonal surface virulence array protein and gene of Aeromonas salmonicida. J Biol Chem 266:15258–15265
Belland RJ, Trust TJ (1987) Cloning of the gene for the surface array protein of Aeromonas salmonicida and evidence linking loss of expression with genetic deletion. J Bacteriol 169:4086–4091. https://doi.org/10.1128/jb.169.9.4086-4091.1987
Silva JB, Storms Z, Sauvageau D (2016) Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363:fnw002. https://doi.org/10.1093/femsle/fnw002
Casjens SR, Molineux IJ (2012) Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Springer, Boston
Lerouge I, Vanderleyden J (2002) O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev 26:17–47. https://doi.org/10.1016/S0168-6445(01)00070-5
Ishiguro EE, Ainsworth T, Shaw DH et al (1983) A lipopolysaccharide-specific bacteriophage for Aeromonas salmonicida. Can J Microbiol 29:1458–1461. https://doi.org/10.1139/m83-223
Lindberg AA (1973) Bacteriophage receptors. Annu Rev Microbiol 27:205–241. https://doi.org/10.1146/annurev.mi.27.100173.001225
González-García VA, Pulido-Cid M, Garcia-Doval C et al (2015) Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J Biol Chem 290:10038–10044. https://doi.org/10.1074/jbc.M114.614222
Rubires X, Merino S, Aguilar A et al (1998) Isolation of three different bacteriophage from mesophilic Aeromonas sp. that use different types of monopolar flagella as their primary receptor. FEMS Microbiol Lett 161:53–57. https://doi.org/10.1016/S0378-1097(98)00049-4
Miñana-Galbis D, Farfán M, Fusté MC, Lorén JG (2004) Aeromonas molluscorum sp. nov., isolated from bivalve molluscs. Int J Syst Evol Microbiol 54:2073–2078. https://doi.org/10.1099/ijs.0.63202-0
Küpfer M, Kuhnert P, Korczak BM et al (2006) Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int J Syst Evol Microbiol 56:2743–2751. https://doi.org/10.1099/ijs.0.63650-0
Burr SE, Pugovkin D, Wahli T et al (2005) Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology 151:2111–2118. https://doi.org/10.1099/mic.0.27926-0
Studer N, Frey J, Vanden Bergh P (2013) Clustering subspecies of Aeromonas salmonicida using IS630 typing. BMC Microbiol 13:36. https://doi.org/10.1186/1471-2180-13-36
Burr SE, Frey J (2007) Analysis of type III effector genes in typical and atypical Aeromonas salmonicida. J Fish Dis 30:711–714. https://doi.org/10.1111/j.1365-2761.2007.00859.x
Massicotte MA, Vincent AT, Schneider A et al (2019) One Aeromonas salmonicida subsp. salmonicida isolate with a pAsa5 variant bearing antibiotic resistance and a pRAS3 variant making a link with a swine pathogen. Sci Total Environ 690:313–320. https://doi.org/10.1016/j.scitotenv.2019.06.456
Vincent AT, Trudel MV, Paquet VE et al (2014) Detection of variants of the pRAS3, pAB5S9, and pSN254 plasmids in Aeromonas salmonicida subsp. salmonicida: Multidrug resistance, interspecies exchanges, and plasmid reshaping. Antimicrob Agents Chemother 58:7367–7374. https://doi.org/10.1128/AAC.03730-14
Nagar V, Shashidhar R, Bandekar JR (2011) Prevalence, characterization, and antimicrobial resistance of Aeromonas strains from various retail food products in Mumbai, India. J Food Sci 76:486–492. https://doi.org/10.1111/j.1750-3841.2011.02303.x
Boutin S, Bernatchez L, Audet C, Derôme N (2012) Antagonistic effect of indigenous skin bacteria of brook charr (Salvelinus fontinalis) against Flavobacterium columnare and F. psychrophilum. Vet Microbiol 155:355–361. https://doi.org/10.1016/j.vetmic.2011.09.002
Benghezal M, Fauvarque MO, Tournebize R et al (2006) Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol 8:139–148. https://doi.org/10.1111/j.1462-5822.2005.00607.x
Berthiaume C, Gilbert Y, Fournier-Larente J et al (2014) Identification of dichloroacetic acid degrading Cupriavidus bacteria in a drinking water distribution network model. J Appl Microbiol 116:208–221. https://doi.org/10.1111/jam.12353
Gauthier J, Charette SJ, Derome N (2017) Draft genome sequence of Pseudomonas fluorescens ML11A, an endogenous strain from brook charr with antagonistic properties against Aeromonas salmonicida subsp. salmonicida. Genome Announc 5:e01716-e1816. https://doi.org/10.1128/genomeA.01716-16
Gonzales MF, Brooks T, Pukatzki SU, Provenzano D (2013) Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J Vis Exp 2013:e50684. https://doi.org/10.3791/50684
Acknowledgements
We would like to thank Sylvain Moineau and Denise Tremblay from the Félix d’Hérelle reference center (U. Laval, Canada). We acknowledge funding from the Ministère de l'agriculture, des pêcheries et de l'alimentation du Québec (INNOVAMER Program), the Natural Sciences and Engineering Research Council of Canada (NSERC) Grant no. RGPIN-2019-04444, and Ressources Aquatiques Québec (RAQ). G.R.L. received an Undergraduate Student Research Award from the NSERC. S.J.C. is a research scholar from the Fonds de Recherche du Québec en Santé (FRQS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing financial interests.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Leduc, G.R., Paquet, V.E., Vincent, A.T. et al. Characterization of bacteriophage T7-Ah reveals its lytic activity against a subset of both mesophilic and psychrophilic Aeromonas salmonicida strains. Arch Virol 166, 521–533 (2021). https://doi.org/10.1007/s00705-020-04923-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04923-w