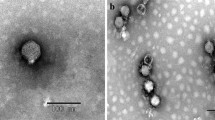
Abstract
A novel Salmonella bacteriophage (phage), named αα, was the first reported member of the family Microviridae to exhibit tolerance to both extreme acidic and alkaline conditions (pH 2-12 for 1 h). Phage αα has a circular single-stranded DNA genome of 5,387 nt with a G+C content of 44.66%. A total of 11 putative gene products and no tRNA genes are encoded in the phage αα genome. Whole-genome sequence comparisons revealed that phage αα shares 95% identity with coliphage phiX174 and had a close evolutionary relationship to the phages NC1 and NC7. Phylogenetic analysis of the structural proteins of phage αα and 18 other phiX174-like phages showed that a phylogenetic tree based on protein B sequences had a topology similar to that obtained using whole genome sequences. In addition, variable sites in proteins F and G distributed on the surface of the mature capsid and the conserved protein J were probably involved in maintaining the structural integrity of the phage under extreme pH conditions. Our findings could open up new perspectives for identifying more extreme-pH-resistant phages and their structural proteins and understanding the mechanism of phage adaptation and evolution under extreme environmental stress.

Similar content being viewed by others
References
Quaiser A, Dufresne A, Ballaud F, Roux S, Zivanovic Y, Colombet J, Sime-Ngando T, Francez AJ (2015) Diversity and comparative genomics of Microviridae in Sphagnum-dominated peatlands. Front Microbiol 6:375
Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV (2018) Viruses of archaea: structural, functional, environmental and evolutionary genomics. Virus Res 244:181–193
Akhwale JK, Rohde M, Rohde C, Bunk B, Spröer C, Boga HI, Klenk H, Wittmann J (2019) Isolation, characterization and analysis of bacteriophages from the haloalkaline lake Elmenteita. Kenya. PLoS One 14:e0215734
Székely AJ, Breitbart M (2016) Single-stranded DNA phages: from early molecular biology tools to recent revolutions in environmental microbiology. FEMS Microbiol Lett 363, fnw027.
Amarillas L, Estrada-Acosta M, León-Chan RG, López-Orona C, Lightbourn L (2020) Complete genome sequence of Phobos: a novel bacteriophage with unusual genomic features that infects Pseudomonas syringae. Arch Virol 165:1485–1488
Mengzhe L, Hong L, Jingxue W, Lei J (2018) Isolation, purification and physiological characteristics of Microviridae bacteriophage αα with a broad pH resistance. Sci Tech Food Ind 2018:15
Rokyta DR, Burch CL, Caudle SB, Wichman HA (2006) Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol 188:1134–1142
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Sayers EW, Beck J, Brister JR, Bolton EE, Canese K, Comeau DC, Funk K, Ketter A, Kim S, Kimchi A, Kitts PA, Kuznetsov A, Lathrop S, Lu Z, McGarvey K, Madden TL, Murphy TD, O’Leary N, Phan L, Schneider VA, Thibaud-Nissen F, Trawick BW, Pruitt KD, Ostell J (2020) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 48:D9–D16
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240
Chan PP, Lowe TM (2019) tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 1962:1–14
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer T, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303
Schrodinger L (2019) The PyMOL molecular graphics system. 2010. Version 1, r1.
Li M, Li M, Lin H, Wang J, Jin Y, Han F (2016) Characterization of the novel T4-like Salmonella enterica bacteriophage STP4-a and its endolysin. Arch Virol 161:377–384
Pavesi A (2006) Origin and evolution of overlapping genes in the family Microviridae. J Gen Virol 87:1013–1017
Yin Y, Ni PE, Deng B, Wang S, Xu W, Wang D (2018) Isolation and characterisation of phages against Pseudomonas syringae pv. actinidiae. Acta Agr Scand. B-S P 69, 199–208
Fogelman I, Davey V, Ochs HD, Elashoff M, Feinberg MB, Mican J, Siegel JP, Sneller M, Lane HC (2000) Evaluation of CD4+ T cell function in vivo in HIV-infected patients as measured by bacteriophage phiX174 immunization. J Infect Dis 182:435–441
Peng W, Si W, Yin L, Liu H, Yu S, Liu S, Wang C, Chang Y, Zhang Z, Hu S, Du Y (2011) Salmonella enteritidis ghost vaccine induces effective protection against lethal challenge in specific-pathogen-free chicks. Immunobiology 216:558–565
Novak CR, Fane BA (2004) The functions of the N terminus of the φX174 internal scaffolding protein, a protein encoded in an overlapping reading frame in a two scaffolding protein system. J Mol Biol 335:383–390
Labrie SJ, Dupuis M, Tremblay DM, Plante P, Corbeil J, Moineau S (2014) A new Microviridae phage isolated from a failed biotechnological process driven by Escherichia coli. Appl Environ Microb 80:6992–7000
Miller CR, Lee KH, Wichman HA, Ytreberg FM (2014) Changing folding and binding stability in a viral coat protein: a comparison between substitutions accessible through mutation and those fixed by natural selection. PLoS ONE 9:e112988
Hafenstein SL, Chen M, Fane BA (2004) Genetic and functional analyses of the φX174 DNA binding protein: the effects of substitutions for amino acid residues that spatially organize the two DNA binding domains. Virology 318:204–213
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31870166 and 32001834) and the Natural Science Foundation of Shandong Province, China (2017GNC13108).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals requiring ethical approval.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, M., Lin, H., Wang, L. et al. Complete genome sequence of the extreme-pH-resistant Salmonella bacteriophage αα of the family Microviridae. Arch Virol 166, 325–329 (2021). https://doi.org/10.1007/s00705-020-04880-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04880-4