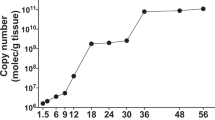
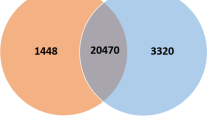
Abstract
Viral hemorrhagic septicemia virus (VHSV) is a rhabdovirus that causes high mortality in cultured flounder. Viral growth and virulence rely on the ability to inhibit the cellular innate immune response. In this study, we investigated differences in the modulation of innate immune responses of HINAE flounder cells infected with low- and high-virulence VHSV strains at a multiplicity of infection of 1 for 12 h and 24 h and performed RNA sequencing (RNA-seq)-based transcriptome analysis. A total of 193 and 170 innate immune response genes were differentially expressed by the two VHSV strains at 12 and 24 h postinfection (hpi), respectively. Of these, 73 and 77 genes showed more than a twofold change in their expression at 12 and 24 hpi, respectively. Of the genes with more than twofold changes, 22 and 11 genes showed high-virulence VHSV specificity at 12 and 24 hpi, respectively. In particular, IL-16 levels were more than two time higher and CCL20a.3, CCR6b, CCL36.1, Casp8L2, CCR7, and Trim46 levels were more than two times lower in high-virulence-VHSV-infected cells than in low-virulence-VHSV-infected cells at both 12 and 24 hpi. Quantitative PCR (qRT-PCR) confirmed the changes in expression of the ten mRNAs with the most significantly altered expression. This is the first study describing the genome-wide analysis of the innate immune response in VHSV-infected flounder cells, and we have identified innate immune response genes that are specific to a high-virulence VHSV strain. The data from this study can contribute to a greater understanding of the molecular basis of VHSV virulence in flounder.







Similar content being viewed by others
References
Walker PJ, Benmansour A, Calisher CH, Dietzgen RG et al (2000) Family rhabdoviridae. Academic Press, San Diego, pp 563–583
Jensen MH (1965) Research on the virus of Egtved disease. Ann NY Acad Sci 126:422–426
Wolf K (1988) Viral hemorrhagic septicemia. Fish viruses and fish viral diseases. Cornell University Press, Ithaca, pp 217–249
Hedrick RP, Batts WN, Yun S, Traxler GS, Kaufman J, Winton JR (2003) Host and geographic range extensions of the North American strain of viral hemorrhagic septicemia virus. Dis Aquat Organ 55:211–220
Takano R, Nishizawa T, Arimoto M, Muroga K (2000) Isolation of viral haemorrhagic septicaemia virus (VHSV) from wild Japanese flounder, Paralichthys olivaceus. Bull Eur Assoc Fish Pathol 20:186–192
Isshiki T, Nishizawa T, Kobayashi T, Nagano T, Miyazaki T (2001) An outbreak of VHSV (viral hemorrhagic septicemia virus) infection in farmed Japanese flounder Paralichthys olivaceus in Japan. Dis Aquat Org 47:87–99
Takano R, Mori K-i, Nishizawa T, Arimoto M, Muroga K (2001) Isolation of viruses from wild Japanese flounder Paralichthys olivaceus. Fish Pathol 36:153–160
Kim S-M, Lee J-I, Hong M-J, Park H-S, Park S-I (2003) Genetic relationship of the VHSV (viral hemorrhagic septicemia virus) isolated from cultured olive flounder, Paralichthys olivaceus in Korea. J Fish Pathol 16:1–12
Kim W-S, Kim S-R, Kim D, Kim J-O, Park M-A, Kitamura S-I, Kim H-Y, Kim D-H, Han H-J, Jung S-J (2009) An outbreak of VHSV (viral hemorrhagic septicemia virus) infection in farmed olive flounder Paralichthys olivaceus in Korea. Aquaculture 296:165–168
McNab F, Mayer-Barber K, Sher A, Wack A, O’garra A (2015) Type I interferons in infectious disease. Nat Rev Immunol 15:87–103
Le CP, Genin P, Baines M, Hiscott J (2000) Interferon activation and innate immunity. Rev Immunogenet 2:374–386
Der SD, Zhou A, Williams BR, Silverman RH (1998) Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA 95:15623–15628
Schneider WM, Chevillotte MD, Rice CM (2014) Interferon-stimulated genes: a complex web of host defenses. Ann Rev Immunol 32:513–545
Rieder M, Conzelmann K-K, Research C (2009) Rhabdovirus evasion of the interferon system. J Interferon Cytokines Res 29:499–510
Haller O, Kochs G, Weber F (2006) The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130
Randall RE, Goodbourn S (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47
Ruiz-Gomez J, Isaacs A (1963) Interferon production by different viruses. Virology 19:8–12
Sellers RF (1963) Multiplication, interferon production and sensitivity of virulent and attenuated strains of the virus of foot-and-mouth disease. Nature 198:1228–1229
Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud J-P, Boudinot P (2013) The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol 425:4904–4920
Zou J, Secombes CJ (2011) Teleost fish interferons and their role in immunity. Dev Comp Immunol 35:1376–1387
Avunje S, Kim W-S, Park C-S, Oh M-J, Jung S-J (2011) Toll-like receptors and interferon associated immune factors in viral haemorrhagic septicaemia virus-infected olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol 31:407–414
Takami IKS, Nishizawa T, Yoshimizu M (2010) Protection of Japanese flounder Paralichthys olivaceus from viral hemorrhagic septicemia (VHS) by Poly(I: C) immunization. Dis Aquat Org 89:109–115
Avunje S, Jung S-J (2017) Poly (I:C) and imiquimod induced immune responses and their effects on the survival of olive flounder (Paralichthys olivaceus) from viral haemorrhagic septicaemia. Fish Shellfish Immunol 71:338–345
Einer-Jensen K, Ahrens P, Forsberg R, Lorenzen N (2004) Evolution of the fish rhabdovirus viral haemorrhagic septicemia virus. J Gen Virol 85:1167–1179
Snow M, Bain N, Black J, Taupin V, Cunningham CO, King JA, Skall HF, Raynard RS (2004) Genetic population structure of marine viral haemorrhagic septicaemia virus (VHSV). Dis Aquat Org 61:11–21
Campbell S, Collet B, Einer-Jensen K, Secombes CJ, Snow M (2009) Identifying potential virulence determinants in viral haemorrhagic septicemia virus (VHSV) for rainbow trout. Dis Aquat Org 86:205–212
Skall HF, Olesen NJ, Mellergaard S (2005) Viral haemorrhagic septicaemia virus in marine fish and its implications for fish farming—a review. J Fish Dis 28:509–529
Skall HF, Slierendrecht WJ, King JA, Olesen NJ (2004) Experimental infection of rainbow trout Oncorhynchus mykiss with viral haemorrhagic septicaemia virus isolates from European marine and farmed fishes. Dis Aquat Org 58:99–110
Emmenegger EJ, Moon CH, Hershberger PK, Kurath G (2013) Virulence of viral hemorrhagic septicemia virus (VHSV) genotypes Ia, IVa, IVb, and IVc in five fish species. Dis Aquat Org 107:99–111
Schönherz AA, Forsberg R, Guldbrandtsen B, Buitenhuis AJ, Einer-Jensen K (2018) Introduction of viral hemorrhagic septicemia virus into freshwater cultured rainbow trout is followed by bursts of adaptive evolution. J Virol 92:e00436-e518
Betts AM, Stone D (2000) Nucleotide sequence analysis of the entire coding regions of virulent and avirulent strains of viral haemorrhagic septicaemia virus. Virus Genes 20:259–262
Kim SH, Thu BJ, Skall HF, Vendramin N, Evensen Ø (2014) A single amino acid mutation (I1012F) of the RNA polymerase of marine viral hemorrhagic septicemia virus changes in vitro virulence to rainbow trout gill epithelial cells. J Virol 88:7189–7198
Einer-Jensen K, Harmache A, Biacchesi S, Bremont M, Stegmann A, Lorenzen N (2014) High virulence differences between phylogenetically distinct isolates of the fish rhabdovirus VHSV is not associated with variability of the surface glycoprotein G nor the nonvirion protein NV. J Gen Virol 95:307–316
Romero A, Figueras A, Tafalla C, Thoulouze MI, Bremont M, Novoa B (2005) Histological, serological and virulence studies on rainbow trout experimentally infected with recombinant infectious hematopoietic necrosis viruses. Dis Aquat Org 68:17–28
Yusuff S, Kurath G, Kim MS, Tesfaye TM, Li J, McKenney DG, Vakharia VN (2019) The glycoprotein, non-virion protein, and polymerase of viral hemorrhagic septicemia virus are not determinants of host-specific virulence in rainbow trout. Virol J 16:31
Byon JY, Ohira T, Hirono I, Aoki T (2005) Use of a cDNA microarray to study immunity against viral hemorrhagic septicemia (VHS) in Japanese flounder (Paralichthys olivaceus) following DNA vaccination. Fish Shellfish Immunol 18:135–147
Byon JY, Ohira T, Hirono I, Aoki T (2006) Comparative immune responses in Japanese flounder, Paralichthys olivaceus after vaccination with viral hemorrhagic septicemia virus (VHSV) recombinant glycoprotein and DNA vaccine using a microarray analysis. Vaccine 24:921–930
Hwang JY, Kwon M-G, Seo JS, Do JW, Park M-A, Jung S-H, Ahn SJ (2016) Differentially expressed genes after viral haemorrhagic septicaemia virus infection in olive flounder (Paralichthys olivaceus). Vet Microbiol 193:72–82
Hwang JY, Kwon MG, Jung S-H, Park MA, Kim D-W, Cho WS, Park JW, Son M-H (2017) RNA-Seq transcriptome analysis of the olive flounder (Paralichthys olivaceus) kidney response to vaccination with heat-inactivated viral hemorrhagic septicemia virus. Fish Shellfish Immunol 62:221–226
Hwang JY, Markkandan K, Kwon MG, Seo JS, Yoo S-i, Hwang SD, Son M-H, Park J (2018) Transcriptome analysis of olive flounder (Paralichthys olivaceus) head kidney infected with moderate and high virulent strains of infectious viral hemorrhagic septicaemia virus (VHSV). Fish Shellfish Immunol 76:293–304
Hwang JY, Markkandan K, Han K, Kwon MG, Seo JS, Yoo S-i, Hwang SD, Ji BY, Son M-H, Park J (2018) Temperature-dependent immune response of olive flounder (Paralichthys olivaceus) infected with viral hemorrhagic septicemia virus (VHSV). Genes Genom 40:315–320
Press CM, Evensen Ø (1999) The morphology of the immune system in teleost fishes. Fish Shellfish Immunol 9:309–318
Kondera E (2014) Cell composition of the head kidney of European chub (Squalius cephalus L.). Arch Pol Fish 22:271–280
Batts W, Winton J (1989) Enhanced detection of infectious hematopoietic necrosis virus and other fish viruses by pretreatment of cell monolayers with polyethylene glycol. J Aquat Anim Health 1:284–290
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Von Mering C (2012) STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815
Park JWMC, Wargo AW, Purcell MK, Kurath G (2010) Differential growth of U and M type infectious hematopoietic necrosis virus in a rainbow trout-derived cell line, RTG-2. J Fish Dis 33:583–591
Wargo AR, Garver KA, Kurath G (2010) Virulence correlates with fitness in vivo for two M group genotypes of Infectious hematopoietic necrosis virus (IHNV). Virology 404:51–58
Wargo AR, Kurath G (2011) In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. J Virol 85:3959–3967
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494
Purcell MK, Kurath G, Garver KA, Herwig RP, Winton J (2004) Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish Immunol 17:447–462
Purcell M, Garver K, Conway C, Elliott D, Kurath G (2009) Infectious haematopoietic necrosis virus genogroup-specific virulence mechanisms in sockeye salmon, Oncorhynchus nerka (Walbaum), from Redfish Lake, Idaho. J Fish Dis 32:619–631
Peñaranda MMDPM, Kurath G (2009) Differential virulence mechanisms of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. J Gen Virol 90:2172–2182
Bernard J, Bearzotti-Le Berre M, De Kinkelin P (1985) Viral haemorrhagic septicaemia in rainbow trout: attempt to relate interferon production, antibody synthesis and structure of the virus with the mechanism of virulence. In: Annales de l'Institut Pasteur/Virologie. Elsevier, pp 13–26
Cho S-Y, Protzman RA, Kim YO, Vaidya B, Oh M-J, Kwon J, Kim D (2019) Elucidation of mechanism for host response to VHSV infection at varying temperatures in vitro and in vivo through proteomic analysis. Fish Shellfish Immunol 88:244–253
Gack MU, Kirchhofer A, Shin YC, Inn K-S, Liang C, Cui S, Myong S, Ha T, Hopfner K-P, Jung JU (2008) Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci USA 105:16743–16748
Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920
Baier M, Werner A, Bannert N, Metzner K, Kurth R (1995) HIV suppression by interleukin-16. Nature 378:563–563
Truong M-J, Darcissac EC, Hermann E, Dewulf J, Capron A, Bahr GM (1999) Interleukin-16 inhibits human immunodeficiency virus type 1 entry and replication in macrophages and in dendritic cells. J Virol 73:7008–7013
Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL (1997) Human CD4+ cells transfected with IL-16 cDNA are resistant to HIV-1 infection: inhibition of mRNA expression. Nat Med 3:659–664
Hu Y, Wei X, Liao Z, Gao Y, Liu X, Su J, Yuan G (2018) Transcriptome analysis provides insights into the markers of resting and LPS-activated macrophages in grass carp (Ctenopharyngodon idella). Int J Mol Sci 19:3562
Wang LZY, Xu WT, Jia XD, Chen SL (2016) Molecular cloning, structure and expressional profiles of two novel single-exon genes (PoCCR6A and PoCCR6B) in the Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol 52:179–188
Liu Y, Chang M, Wu S, Nie P (2009) Characterization of C-C chemokine receptor subfamily in teleost fish. Mol Immunol 46:498–504
Schutyser E, Struyf S, Van Damme J (2003) The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev 14:409–426
Kallal LE, Schaller MA, Lindell DM, Lira SA, Lukacs NW (2010) CCL20/CCR6 blockade enhances immunity to RSV by impairing recruitment of DC. Eur J Immunol 40:1042–1052
Lee AY, Körner H (2017) CCR6/CCL20 chemokine axis in human immunodeficiency virus immunity and pathogenesis. J Gen Virol 98:338–344
Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ (2001) Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci USA 98:13884–13888
Cheng F, Chen AY, Best SM, Bloom ME, Pintel D, Qiu J (2010) The capsid proteins of Aleutian mink disease virus activate caspases and are specifically cleaved during infection. J Virol 84:2687–2696
Best SM, Shelton JF, Pompey JM, Wolfinbarger JB, Bloom ME (2003) Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. J Virol 77:5305–5312
Förster R, Pabst O, Bernhardt G (2008) Homeostatic chemokines in development, plasticity, and functional organization of the intestinal immune system. In: Seminars in immunology. Elsevier, pp 171–180
Kocks JR, Adler H, Danzer H, Hoffmann K, Jonigk D, Lehmann U, Förster R (2009) Chemokine receptor CCR7 contributes to a rapid and efficient clearance of lytic murine γ-herpes virus 68 from the lung, whereas bronchus-associated lymphoid tissue harbors virus during latency. J Immunol 182:6861–6869
Bardina SV, Brown JA, Michlmayr D, Hoffman KW, Sum J, Pletnev AG, Lira SA, Lim JK (2017) Chemokine receptor Ccr7 restricts fatal West Nile virus encephalitis. J Virol 91:e02409-02416
Ordás MC, Castro R, Dixon B, Sunyer JO, Bjork S, Bartholomew J, Korytar T, Köllner B, Cuesta A, Tafalla C (2012) Identification of a novel CCR7 gene in rainbow trout with differential expression in the context of mucosal or systemic infection. Dev Comp Immunol 38:302–311
Aquilino C, Castro R, Fischer U, Tafalla C (2014) Transcriptomic responses in rainbow trout gills upon infection with viral hemorrhagic septicemia virus (VHSV). Dev Comp Immunol 44:12–20
Short KM, Cox TC (2006) Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem 281:8970–8980
Ichinose S, Ogawa T, Jiang X, Hirokawa N (2019) The spatiotemporal construction of the axon initial segment via KIF3/KAP3/TRIM46 transport under MARK2 signaling. Cell Rep 28(2413–2426):e2417
van Beuningen SF, Will L, Harterink M, Chazeau A et al (2015) TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 88:1208–1226
Acknowledgements
This work was supported by a grant from the National Institute of Fisheries Science in the Republic of Korea (Grant Number: R2019058) and the National Research Foundation of the Republic of Korea (2019R1A2C1006721).
Author information
Authors and Affiliations
Contributions
JYH collected VHSV strains and performed the in vivo infection experiment. UHL performed the in vitro VHSV growth experiment. JYH and UHL analyzed RNA-seq data. MJH and JMJ performed the in vivo infection experiment and analyzed RNA-seq data. MGK and BYJ designed the experiments and discussed the results. CIP and JWP designed the experiments, wrote the manuscript, and analyzed the results.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing financial interests.
Additional information
Handling Editor: William G Dundon.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2020_4871_MOESM1_ESM.xlsx
Supplementary file1 Supplemental Table S1 List of DEGs between mock-infected control and VHS2012-6-infected HINAE cells at 12 h postinfection (XLSX 6674 KB)
705_2020_4871_MOESM2_ESM.xlsx
Supplementary file2 Supplemental Table S2 List of DEGs between mock-infected control and VHS2015-5-infected HINAE cells at 12 h postinfection (XLSX 6730 KB)
705_2020_4871_MOESM3_ESM.xlsx
Supplementary file3 Supplemental Table S3 List of DEGs between mock-infected control and VHS2012-6-infected HINAE cells at 24 h postinfection (XLSX 6636 KB)
705_2020_4871_MOESM4_ESM.xlsx
Supplementary file4 Supplemental Table S4 List of DEGs between mock-infected control and VHS2015-5-infected HINAE cells at 24 h postinfection (XLSX 6571 KB)
705_2020_4871_MOESM5_ESM.xlsx
Supplementary file5 Supplemental Table S5 List of innate immune response genes differentially expressed in VHS2015-5-infected and VHS2012-6-infected HINAE cells compared to mock-infected control HINAE cells at 12 h postinfection (XLSX 19 KB)
705_2020_4871_MOESM6_ESM.xlsx
Supplementary file6 Supplemental Table S6 List of innate immune response genes differentially expressed in VHS2015-5-infected and VHS2012-6-infected HINAE cells compared to mock-infected control HINAE cells at 24 h postinfection (XLSX 18 KB)
Rights and permissions
About this article
Cite this article
Hwang, J.Y., Lee, U.H., Heo, M.J. et al. RNA-seq transcriptome analysis in flounder cells to compare innate immune responses to low- and high-virulence viral hemorrhagic septicemia virus. Arch Virol 166, 191–206 (2021). https://doi.org/10.1007/s00705-020-04871-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04871-5