Abstract
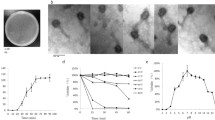
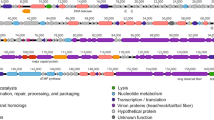
Enterobacter aerogenes is a member of the ESKAPE group of bacteria, and multi-drug-resistant strains are increasingly being found. In this study, a novel bacteriophage, ATCEA85, which infects E. aerogenes, has been isolated and characterized. ATCEA85 is seen to have a circularly permuted linear double-stranded DNA genome of 47,484 base pairs in length. The closest related phage found in the databases is the Klebsiella phage Kp3, which exhibits 77% identity over a 34% query coverage. The G+C content of ATCEA85 is 56.2%, and 15 putative open reading frames are functionally annotated.


Similar content being viewed by others
References
Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR (2019) Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539
Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308
Davin-Regli A, Pagès JM (2019) Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392
Davin-Regli A, Lavigne JP, Pagès JM (2019) Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev 32(4):e00002–19. https://doi.org/10.1128/CMR.00002-19
Thaden JT, Pogue JM, Kaye KS (2017) Role of newer and re-emerging older agents in the treatment of infections caused by carbapenem-resistant Enterobacteriaceae. Virulence 8(4):403–416
Rozwandowicz M, Brouwer MSM, Fischer J et al (2018) Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73(5):1121–1137
Fritzenwanker M, Imirzalioglu C, Herold S et al (2018) Treatment options for carbapenem-resistant Gram-negative infections. Dtsch Arztebl Int. 115(20–21):345–352
Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R (2019) Therapeutic characterization and Efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol 10:574
Verthé K, Possemiers S, Boon N, Vaneechoutte M, Verstraete W (2004) Stability and activity of an Enterobacter aerogenes-specific bacteriophage under simulated gastro-intestinal conditions. Appl Microbiol Biotechnol 65(4):465–472
Mishra CK, Choi TJ, Kang SC (2012) Isolation and characterization of a bacteriophage F20 virulent to Enterobacter aerogenes. J Gen Virol 93(Pt 10):2310–2314
Li E, Wei X, Ma Y, Yin Z, Li H, Lin W, Wang X, Li C, Shen Z, Zhao R, Yang H, Jiang A, Yang W, Yuan J, Zhao X (2016) Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci Rep 6:28338
Zhao J, Zhang Z, Tian C, Chen X, Hu L, Wei X, Li H, Lin W, Jiang A, Feng R, Yuan J, Yin Z, Zhao X (2019) Characterizing the biology of lytic bacteriophage vB_EaeM_φEap-3 infecting multidrug-resistant Enterobacter aerogenes. Front Microbiol 10:420
Domingo-Calap P, Delgado-Martínez J (2018) Bacteriophages: protagonists of a post-antibiotic era. Antibiotics 7(3):66
Hong SS, Jeong J, Lee J, Kim S, Min W, Myung H (2013) Therapeutic effects of bacteriophages against Salmonella gallinarum infection in chickens. J Microbiol Biotechnol 23:1478–1483
Cha K, Oh HK, Jang JY, Jo Y, Kim WK, Ha GU, Ko KS, Myung H (2018) Characterization of two novel bacteriophages infecting multidrug-resistant (MDR) Acinetobacter baumannii and evaluation of their therapeutic efficacy in vivo. Front Microbiol 9:696
Azam AH, Tanji Y (2019) Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol 103(5):2121–2131
Divya Ganeshan S, Hosseinidoust Z (2019) Phage therapy with a focus on the human microbiota. Antibiotics 8(3):131
Bernheim A, Sorek R (2019) The pan-immune system of bacteria: antiviral defence as a community resource. Nat Rev Microbiol 18:113–119
Sambrook J, Russell DW (2006) Purification of bacteriophage lambda particles by centrifugation through a glycerol step gradient. CSH Protoc. https://doi.org/10.1101/pdb.prot3969
Manfioletti G, Schneider C (1988) A new and fast method for preparing high quality lambda DNA suitable for sequencing. Nucleic Acids Res 16:2873–2884
Garneau JR, Depardieu F, Fortier L, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7(1):1–10
Garneau JR, Sekulovic O, Dupuy B, Soutourina O, Monot M, Fortier LC (2018) High prevalence and genetic diversity of large phiCD211 (phiCDIF1296T)-like prophages in Clostridioides difficile. Appl Environ Microbiol 84(3) (pii: e02164-17)
Funding
This work was supported by the ATC Program 10076996 from Ministry of Trade, Industry, and Energy of Korea. In addition, this study was sponsored by the National Research Foundation of Korea (NRF-2017M3A9B8069292) and the HUFS Research Fund of 2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This study does not contain any experiments with human or animal subjects performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Oh, H.K., Jo, J.H., Hwang, Y.J. et al. Complete genome sequence of a novel bacteriophage, ATCEA85, infecting Enterobacter aerogenes. Arch Virol 165, 2397–2400 (2020). https://doi.org/10.1007/s00705-020-04751-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04751-y