Abstract
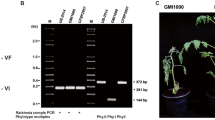
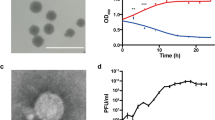
Bacterial wilt caused by Ralstonia spp., soil-borne Gram-negative bacteria, is considered one of the most important plant diseases in tropical and subtropical regions of the world. A large number of bacteriophages capable of lysing or physiologically reprogramming cells of Ralstonia spp. have been reported in Asia. Despite the potential use of these organisms in the management of bacterial wilt, information on viruses that infect Ralstonia spp. is nonexistent in the Americas. We isolated a virus that infects Ralstonia spp. from a soil sample in the state of Minas Gerais, Brazil. Microscopy and genomic and phylogenetic analysis allowed us to classify the virus as a member of the family Podoviridae, genus Phikmvvirus. In spite of its relationship to Ralstonia virus RSB3, an Asian isolate, genomic and biological characteristics showed that the virus isolated in Brazil, tentatively named “Ralstonia virus phiAP1” (phiAP1), belongs to a new species. phiAP1 has EPS depolymerase activity and contains two putative virion-associated peptidoglycan hydrolases (VAPGHs), which reveals a robust mechanism of pathogenesis. Furthermore, phiAP1 specifically infects Ralstonia solanacearum, R. pseudosolanacearum and R. syzygii, causing cell lysis, but it was not able to infect thirteen other bacteria that were tested. Together, these characteristics highlight the biotechnological potential of this virus for the management of bacterial wilt.







Similar content being viewed by others
References
Cassman N, Prieto-Davó A, Walsh K et al (2012) Oxygen minimum zones harbour novel viral communities with low diversity. Environ Microbiol 14:3043–3065. https://doi.org/10.1111/j.1462-2920.2012.02891.x
Dutilh BE (2014) Metagenomic ventures into outer sequence space. Bacteriophage 4:3–5. https://doi.org/10.4161/21597081.2014.979664
Klumpp J, Fouts DE, Sozhamannan S (2012) Next generation sequencing technologies and the changing landscape of phage genomics. Bacteriophage 2:190–199. https://doi.org/10.4161/bact.22111
Balogh B, Jones JB, Iriarte FB, Momol MT (2010) Phage therapy for plant disease control. Curr Pharm Biotechnol 11:48–57. https://doi.org/10.2174/138920110790725302
Frampton RA, Pitman AR, Fineran PC (2012) Advances in bacteriophage-mediated control of plant pathogens. Int J Microbiol 2012:1–11. https://doi.org/10.1155/2012/326452
Oliveira H, Sillankorva S, Merabishvili M et al (2015) Unexploited opportunities for phage therapy. Front Pharmacol 6:1–4. https://doi.org/10.3389/fphar.2015.00180
Álvarez B, Biosca EG, López MM (2010) On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. Technol Educ Top Appl Microbiol Microb Biotechnol 1:267–279
Mansfield J, Genin S, Magori S et al (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. https://doi.org/10.1111/j.1364-3703.2012.00804.x
Prior P, Ailloud F, Dalsing BL et al (2016) Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genom 17:90. https://doi.org/10.1186/s12864-016-2413-z
Safni I, Cleenwerck I, De Vos P et al (2014) Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii. Int J Syst Evol Microbiol 64:3087–3103. https://doi.org/10.1099/ijs.0.066712-0
Hayward AC (1991) Biology and edidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29:65–87
Yuliar Nion YA, Toyota K (2015) Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ 30:1–11. https://doi.org/10.1264/jsme2.ME14144
Allen C, Prior P, Hayward A (2005) Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, Saint Paul, USA
Wang JF, Lin CH (2005) Colonization capacity of Ralstonia solanacearum tomato strains differing in aggressiveness on tomatoes and weeds. In: Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, Saint Paul, USA, pp 73–79
Toyoda H, Kakutani K, Ikeda S et al (1991) Characterization of deoxyribonucleic acid of virulent bacteriophage and its infectivity to host bacteria, Pseudomonas solanacearum. J Phytopathol 131:11–21. https://doi.org/10.1111/j.1439-0434.1991.tb04566.x
Ozawa H, Tanaka H, Ichinose Y et al (2001) Bacteriophage P4282, a parasite of Ralstonia solanacearum, encodes a bacteriolytic protein important for lytic infection of its host. Mol Genet Genom 265:95–101. https://doi.org/10.1007/s004380000389
Kawasaki T, Narulita E, Matsunami M et al (2016) Genomic diversity of large-plaque-forming podoviruses infecting the phytopathogen Ralstonia solanacearum. Virology 492:73–81. https://doi.org/10.1016/j.virol.2016.02.011
Kawasaki T, Shimizu M, Satsuma H et al (2009) Genomic characterization of Ralstonia solanacearum phage φRSB1, a T7-like wide-host-range phage. J Bacteriol 91:422–427. https://doi.org/10.1128/JB.01263-08
Addy HS, Askora A, Kawasaki T et al (2012) Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by φRSM filamentous phages. Phytopathology 102:469–477. https://doi.org/10.1094/PHYTO-11-11-0319-R
Yamada T (2012) Bacteriophages of Ralstonia solanacearum: their diversity and utilization as biocontrol agents in agriculture. In: Kurtboke I (ed) Bacteriophages. InTech China, Shanghai, pp. 113–138. http://www.intechopen.com/books/bacteriophages/bacteriophages-of-ralstonia-solanacearum-their-diversity-and-utilization-as-biocontrol-agents-in-agr
Van Truong Thi B, Pham Khanh NH, Namikawa R et al (2016) Genomic characterization of Ralstonia solanacearum phage ϕRS138 of the family Siphoviridae. Arch Virol 161:483–486. https://doi.org/10.1007/s00705-015-2654-1
Adams MH (1959) Bacteriophages. Interscience Publishers, New York
Yamada T, Kawasaki T, Nagata S et al (2007) New bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 153:2630–2639. https://doi.org/10.1099/mic.0.2006/001453-0
Sambrook J, Russel D (2001) Molecular cloning—a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST:a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Besemer J, Borodovsky M (1999) Heuristic approach to deriving models for gene finding. Nucleic Acids Res 27:3911–3920. https://doi.org/10.1093/nar/27.19.3911
Hyatt D, Chen G-L, Locascio PF et al (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11:119. https://doi.org/10.1186/1471-2105-11-119
Krogh A, Larsson B, von Heijne G, Sonnhammer E (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Käll L, Krogh A, Sonnhammer ELL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. https://doi.org/10.1016/j.jmb.2004.03.016
Kelley LA, Sternberg MJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. https://doi.org/10.1038/nprot.2009.2
Marchler-Bauer A, Zheng C, Chitsaz F et al (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:1–5. https://doi.org/10.1093/nar/gks1243
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56. https://doi.org/10.1016/S0097-8485(01)00099-7
Gautheret D, Lambert A (2001) Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol 313:1003–1011. https://doi.org/10.1006/jmbi.2001.5102
Macke TJ, Ecker DJ, Gutell RR et al (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29:4724–4735. https://doi.org/10.1093/nar/29.22.4724
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Darling AE, Mau B, Perna NT (2010) Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Kropinski AM, Prangishvili D, Lavigne R (2009) Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ Microbiol 11:2775–2777. https://doi.org/10.1111/j.1462-2920.2009.01970.x
Mahadevan P, Seto D (2009) Data mining pathogen genomes using GeneOrder and CoreGenes: gene order, synteny, and proteomes. Int J Comput Biol Drug Des 2:100–114
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Krumsiek J, Arnold R, Rattei T (2007) Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. https://doi.org/10.1093/bioinformatics/btm039
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. https://doi.org/10.1371/journal.pone.0108277
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67
Moak M, Molineux IJ (2004) Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol 51:1169–1183. https://doi.org/10.1046/j.1365-2958.2003.03894.x
Welker P, Geist B, Fruhauf JH et al (2007) Role of lipid rafts in membrane delivery of renal epithelial Na+-K+-ATPase, thick ascending limb. Am J Physiol Regul Integr Comp Physiol 292:R1328–R1337. https://doi.org/10.1152/ajpregu.00166.2006
Pantůček R, Rosypalová A, Doškař J et al (2008) The polyvalent staphylococcal phage phi812: its host-range mutants and related phages. Virology 246:241–252. https://doi.org/10.1006/viro.1998.9203
Ceyssens PJ, Lavigne R, Mattheus W et al (2006) Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the φKMV subgroup within the T7 supergroup. J Bacteriol 188:6924–6931. https://doi.org/10.1128/JB.00831-06
Lavigne R, Burkal’tseva MV, Robben J et al (2003) The genome of bacteriophage φKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 312:49–59. https://doi.org/10.1016/S0042-6822(03)00123-5
Yu X, Xu Y, Gu Y et al (2017) Characterization and genomic study of “phiKMV-Like” phage PAXYB1 infecting Pseudomonas aeruginosa. Sci Rep 7:13068. https://doi.org/10.1038/s41598-017-13363-7
Gill JJ, Summer EJ, Russell WK et al (2011) Genomes and characterization of phages Bcep22 and BcepIL02, founders of a novel phage type in Burkholderia cenocepacia. J Bacteriol 193:5300–5313. https://doi.org/10.1128/JB.05287-11
Yamada T, Satoh S, Ishikawa H et al (2010) A jumbo phage infecting the phytopathogen Ralstonia solanacearum defines a new lineage of the Myoviridae family. Virology 398:135–147. https://doi.org/10.1016/j.virol.2009.11.043
Adriaenssens EM, Ceyssens PJ, Dunon V et al (2011) Bacteriophages LIMElight and LIMEzero of Pantoea agglomerans, belonging to the “phiKMV-Like Viruses”. Appl Environ Microbiol 77:3443–3450. https://doi.org/10.1128/AEM.00128-11
Ahern SJ, Das M, Bhowmick TS et al (2014) Characterization of novel virulent broad-host-range phages of Xylella fastidiosa and Xanthomonas. J Bacteriol 196:459–471. https://doi.org/10.1128/JB.01080-13
Kulakov LA, Ksenzenko VN, Shlyapnikov MG et al (2009) Genomes of “phiKMV-like viruses” of Pseudomonas aeruginosa contain localized single-strand interruptions. Virology 391:1–4. https://doi.org/10.1016/j.virol.2009.06.024
Lammens E, Ceyssens P-J, Voet M et al (2009) Representational difference analysis (RDA) of bacteriophage genomes. J Microbiol Methods 77:207–213. https://doi.org/10.1016/j.mimet.2009.02.006
Lynch KH, Abdu AH, Schobert M, Dennis JJ (2013) Genomic characterization of JG068, a novel virulent podovirus active against Burkholderia cenocepacia. BMC Genom 14:574. https://doi.org/10.1186/1471-2164-14-574
Kawasaki T, Shimizu M, Satsuma H et al (2009) Genomic characterization of Ralstonia solanacearum phage RSB1, a T7-like wide-host-range phage. J Bacteriol 191:422–427. https://doi.org/10.1128/JB.01263-08
Adams MJ, King AMQ, Carstens EB (2013) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013). Arch Virol 158:2023–2030. https://doi.org/10.1007/s00705-013-1688-5
Carstens EB (2010) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009). Arch Virol 155:133–146. https://doi.org/10.1007/s00705-009-0547-x
Lai MJ, Chang KC, Huang SW et al (2016) The tail associated protein of Acinetobacter baumannii phage qab6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS One 11:1–14. https://doi.org/10.1371/journal.pone.0153361
Olszak T, Shneider MM, Latka A et al (2017) The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep 7:1–14. https://doi.org/10.1038/s41598-017-16411-4
Ailloud F, Lowe T, Cellier G et al (2015) Comparative genomic analysis of Ralstonia solanacearum reveals candidate genes for host specificity. BMC Genom 16:270. https://doi.org/10.1186/s12864-015-1474-8
Roach DR, Sjaarda DR, Castle AJ, Svircev AM (2013) Host exopolysaccharide quantity and composition impact Erwinia amylovora bacteriophage pathogenesis. Appl Environ Microbiol 79:3249–3256. https://doi.org/10.1128/AEM.00067-13
Scholl D, Adhya S, Merril C (2005) Escherichia coli K1’s capsule is a barrier to bacteriophage T7. Appl Environ Microbiol 71:4872–4874. https://doi.org/10.1128/AEM.71.8.4872
Denny TP (1995) Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol 33:173–197. https://doi.org/10.1146/annurev.py.33.090195.001133
Genin S, Boucher CA (2002) Ralstonia solanacearum: secretes of a major pathogen unveiled by analysis of its genome. Mol Plant Pathol 3:111–118
Malnoy MM, Faize M, Venisse J-SJ-S et al (2005) Expression of viral EPS-depolymerase reduces fire blight susceptibility in transgenic pear. Plant Cell Rep 23:632–638. https://doi.org/10.1007/s00299-004-0855-2
Kim WS, Salm H, Geider K (2004) Expression of bacteriophage φEa1h lysozyme in Escherichia coli and its activity in growth inhibition of Erwinia amylovora. Microbiology 150:2707–2714. https://doi.org/10.1099/mic.0.27224-0
Zhang Y, Goritschnig S, Dong X, Li X (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15:2636–2646. https://doi.org/10.1105/tpc.015842.nb-lrr
Rodríguez-Rubio L, Martínez B, Donovan DM et al (2013) Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. Crit Rev Microbiol 39:427–434. https://doi.org/10.3109/1040841X.2012.723675
Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B (2015) Bacteriophages and phage-derived proteins—application approaches. Curr Med Chem 22:1757–1773. https://doi.org/10.2174/0929867322666150209152851
Evanics F, Hwang PM, Cheng Y et al (2006) Topology of an outer-membrane enzyme: measuring oxygen and water contacts in solution NMR studies of PagP. J Am Chem Soc 128:8256–8264. https://doi.org/10.1021/ja0610075
Jia W, El Zoeiby A, Petruzziello TN et al (2004) Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem 279:44966–44975. https://doi.org/10.1074/jbc.M404963200
Murata T, Tseng W, Guina T et al (2007) PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol 189:7213–7222. https://doi.org/10.1128/JB.00973-07
Barbosa TM, Levy SB (2000) Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 182:3467–3474. https://doi.org/10.1128/jb.182.12.3467-3474.2000.updated
Briers Y, Peeters LM, Volckaert G, Lavigne R (2011) The lysis cassette of bacteriophage ϕKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage 1:25–30. https://doi.org/10.4161/bact.1.1.14868
Summer EJ, Berry J, Tran TAT et al (2007) Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J Mol Biol 373:1098–1112. https://doi.org/10.1016/j.jmb.2007.08.045
Acknowledgements
We thank F. Murilo Zerbini and the members of our laboratory for helpful discussions, Karla Ribeiro and Gilmar Valente (Núcleo de Microscopia e Microanálise, CCB/UFV) for assistance with TEM analysis, and Edvaldo Barros (Núcleo de Análise de Biomoléculas, CCB/UFV) for assistance with proteomic analysis. We also thank Rosa de Lima Ramos Mariano and Elineide Barbosa de Souza (Dep. de Fitossanidade, UFRPE), Carlos Alberto Lopes (EMBRAPA-CNPH), Valmir Duarte (Agronômica Laboratório de Diagnóstico Fitossanitário), Acelino Couto Alfenas and José Rogério de Oliveira (Dep. de Fitopatologia, UFV), Caitilyn Allen (University of Wisconsin-Madison) and Rosalee Albuquerque Coelho Neto (Instituto Nacional de Pesquisa da Amazônia) for providing the bacterial isolates used in this work. ASX was the recipient of a FAPEMIG graduate fellowship, TTML was the recipient of a CNPq undergraduate fellowship, FOS and FPS were the recipients of CNPq graduate fellowships.
Funding
This study was funded by Grant APQ-01926-14 (FAPEMIG) to PAZ.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animal performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
da Silva Xavier, A., da Silva, F.P., Vidigal, P.M.P. et al. Genomic and biological characterization of a new member of the genus Phikmvvirus infecting phytopathogenic Ralstonia bacteria. Arch Virol 163, 3275–3290 (2018). https://doi.org/10.1007/s00705-018-4006-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4006-4