Abstract
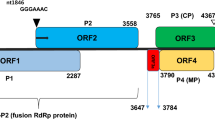
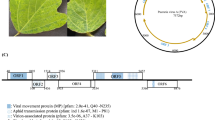
We determined the complete genome sequence of a putative novel ilarvirus, tentatively named “peanut virus C” (PVC), identified in peanut (Arachis hypogaea). The three segmented genomic RNA molecules of PVC were 3474 (RNA1), 2925 (RNA2), and 2160 (RNA3) nucleotides in length, with five predicted open reading frames containing conserved domains and motifs that are typical features of ilarviruses. The three genomic RNAs shared nucleotide sequence similarity (74% identity and 93% query coverage for RNA1, 75% identity and 85% query coverage for RNA2, and 72% identity and 70% query coverage for RNA3) with the most closely related ilarvirus, parietaria mottle virus. These results suggest that PVC is a novel member of the genus Ilarvirus in the family Bromoviridae.


Similar content being viewed by others
References
Adams IP, Glover RH, Monger WA, Mumford R, Jackeviciene E, Navalinskiene M, Samuitiene M, Boonham N (2009) Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol Plant Pathol 10(4):537–545
Barba M, Czosnek H, Hadidi A (2014) Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 6(1):106–136
Boonham N, Kreuze J, Winter S, Vlugt RVD, Bergervoet J, Tomlinson J, Mumford R (2014) Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res 186:20–31
Brister JR, Ako-Adjei D, Bao Y, Blinkova O (2015) NCBI viral genomes resource. Nucleic Acids Res 43(Database issue):D571–D577
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42(Database issue):D222–D230
James D, Varga A (2012) Sequence analysis of RNA 1 of lilac leaf chlorosis virus supports a close relationship to subgroup 3 ilarviruses. Arch Virol 157(1):203–206
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lim S, Kim KH, Zhao F, Yoo RH, Igori D, Lee SH, Moon JS (2015) Complete genome sequence of a novel endornavirus isolated from hot pepper. Arch Virol 160(12):3153–3156
Pallas V, Aparicio F, Herranz MC, Amari K, Sanchez-Pina MA, Myrta A, Sanchez-Navarro JA (2012) Ilarviruses of Prunus spp.: a continued concern for fruit trees. Phytopathology 102(12):1108–1120
Pallas V, Aparicio F, Herranz MC, Sanchez-Navarro JA, Scott SW (2013) The molecular biology of ilarviruses. Adv Virus Res 87:139–181
Scott SW, Zimmerman MT (2006) The complete sequence of the genome of Humulus japonicus latent virus. Arch Virol 151(8):1683–1687
Acknowledgments
This research was carried out with the support of the Cooperative Research Program for Agriculture Science and Technology Development (project no. PJ01248602), Rural Development Administration, Republic of Korea. We thank Ms. M. H. Jung for her technical assistance, and Sarah Williams, PhD, from Edanz Group (www.edanzediting.com) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ioannis E. Tzanetakis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lim, S., Jang, YW., Bae, J.W. et al. Complete genome sequence of peanut virus C, a putative novel ilarvirus. Arch Virol 163, 2265–2269 (2018). https://doi.org/10.1007/s00705-018-3827-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3827-5