Abstract
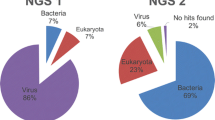
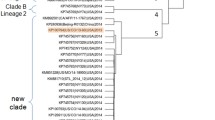
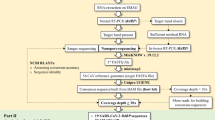
Metagenomic analysis through high-throughput sequencing is a tool for detecting both known and novel viruses. Using this technique, a novel circular single-stranded DNA (ssDNA) virus genome was discovered in respiratory secretions from a febrile traveler. The virus, named human respiratory-associated PSCV-5-like virus (HRAPLV), has a genome comprising 3,018 bases, with two major putative ORFs inversely encoding capsid (Cap) and replicase (Rep) protein and separated by two intergenic regions. One stem-loop structure was predicted in the larger intergenic region (LIR). The predicted amino acid sequences of the Cap and Rep proteins of HRAPLV showed highest identity to those of porcine stool-associated circular virus 5 isolate CP3 (PoSCV 5) (53.0% and 48.9%, respectively). The host tropism of the virus is unknown, and further study is warranted to determine whether this novel virus is associated with human disease.




Similar content being viewed by others
References
Blinkova O, Victoria J, Li Y, Keele BF, Sanz C, Ndjango JB, Peeters M, Travis D, Lonsdorf EV, Wilson ML, Pusey AE, Hahn BH, Delwart EL (2010) Novel circular DNA viruses in stool samples of wild-living chimpanzees. J Gen Virol 91:74–86
Cheung AK, Ng TF, Lager KM, Alt DP, Delwart EL, Pogranichniy RM (2014) Identification of a novel single-stranded circular DNA virus in pig feces. Genome Announc 2:e00347-14
Daly GM, Bexfield N, Heaney J, Stubbs S, Mayer AP, Palser A, Kellam P, Drou N, Caccamo M, Tiley L, Alexander GJ, Bernal W, Heeney JL (2011) A viral discovery methodology for clinical biopsy samples utilising massively parallel next generation sequencing. PloS One 6:e28879
Delwart E, Li L (2012) Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res 164:114–121
Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D (2007) Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 3:e64
Halary S, Duraisamy R, Fancello L, Monteil-Bouchard S, Jardot P, Biagini P, Gouriet F, Raoult D, Desnues C (2016) Novel single-stranded DNA circular viruses in pericardial fluid of patient with recurrent pericarditis. Emerg Infect Dis 22:1839–1841
Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS (2003) Unbiased whole-genome amplification directly from clinical samples. Genome Res 13:954–964
Huson DH, Bryant D (2006) Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol 23:254–267
Kim HK, Park SJ, Nguyen VG, Song DS, Moon HJ, Kang BK, Park BK (2012) Identification of a novel single-stranded, circular DNA virus from bovine stool. The J Gen Virol 93:635–639
Lamberto I, Gunst K, Muller H, Zur Hausen H, de Villiers EM (2014) Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc 2:e00848-14
Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E (2010) Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84:1674–1682
Liu S, Xie J, Cheng J, Li B, Chen T, Fu Y, Li G, Wang M, Jin H, Wan H, Jiang D (2016) Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc Natl Acad Sci USA 113:12803–12808
Lopez-Bueno A, Rastrojo A, Peiro R, Arenas M, Alcami A (2015) Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Mol Ecol 24:4812–4825
Phan TG, Luchsinger V, Avendano LF, Deng X, Delwart E (2014) Cyclovirus in nasopharyngeal aspirates of Chilean children with respiratory infections. J Gen Virol 95:922–927
Rosario K, Duffy S, Breitbart M (2012) A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol 157:1851–1871
Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM (2017) Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168
Smits SL, Zijlstra EE, van Hellemond JJ, Schapendonk CM, Bodewes R, Schurch AC, Haagmans BL, Osterhaus AD (2013) Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010–2011. Emerg Infect Dis 19:1511–1513
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
le Tan V, van Doorn HR, Nghia HD, Chau TT, le Tu TP, de Vries M, Canuti M, Deijs M, Jebbink MF, Baker S, Bryant JE, Tham NT, BKong NT, Boni MF, Loi TQ, le Phuong T, Verhoeven JT, Crusat M, Jeeninga RE, Schultsz C, Chau NV, Hien TT, van der Hoek L, Farrar J, de Jong MD (2013) Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. mBio 4:e00231-13
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X (2010) A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA 107:8387–8392
Zhou C, Zhang S, Gong Q, Hao A (2015) A novel gemycircularvirus in an unexplained case of child encephalitis. Virol J 12:197
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This study was funded in part by Science and Technology Support Program of Jiangsu Province for Emerging Infectious Diseases Control and Prevention (No. BE2015714), Key Medical Discipline of Jiangsu Province (ZDXKA2016008), the National Natural Science Foundation of China (81501785), the Natural Science Foundation of Jiangsu Province (BK20141030, BK20161583), and the “333” Projects of Jiangsu Province.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of Jiangsu Provincial Center for Disease Control and Prevention and Jiangsu International Travel Healthcare Center.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3481_MOESM1_ESM.tif
Fig. S1 NeighborNet networks based on the complete genome sequence of HRAPLV and other circular single-stranded DNA viruses. A filled diamond indicates the sample obtained in this study (TIFF 274 kb)
Rights and permissions
About this article
Cite this article
Cui, L., Wu, B., Zhu, X. et al. Identification and genetic characterization of a novel circular single-stranded DNA virus in a human upper respiratory tract sample. Arch Virol 162, 3305–3312 (2017). https://doi.org/10.1007/s00705-017-3481-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3481-3