Abstract
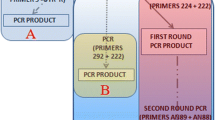
Recently, a reverse transcriptase semi-nested polymerase chain reaction (RT-snPCR) assay was recommended by the WHO for direct detection of enteroviruses in clinical specimens. In this study, we use this assay and a modification thereof to screen acute flaccid paralysis (AFP) samples that had previously tested negative for enteroviruses by the RD-L20B algorithm. Thirty paired stool suspensions collected in 2015 as part of the national AFP surveillance program in different states of Nigeria were analyzed in this study. The samples had previously tested negative for enteroviruses in the polio laboratory in accordance with the WHO-recommended RD-L20B-cell-culture-based algorithm. Two samples that had previously been found to contain enteroviruses were included as positive controls. All samples were subjected to RNA extraction, the RT-snPCR assay and a modified version of the RT-snPCR. All amplicons were sequenced, and enteroviruses were identified using the enterovirus genotyping tool and phylogenetic analysis. Amplicons were recovered from the two controls and 50% (15/30) of the samples screened. Fourteen were successfully typed, of which, 7.1% (1/14), 21.4% (3/14), 64.3% (9/14) and 7.1% (1/14) were enterovirus (EV) -A, EV-B, EV-C and a mixture of EV-B and C (EV-C99 and E25), respectively. The two controls were identified as EV-C99 and coxsackievirus (CV) -A1, both of which belong to the species Enterovirus C. In one sample, poliovirus serotype 2 was detected and found to have the VP1 ILE143 variation and was therefore identified as a vaccine strain. The results of this study showed that significant proportion of enterovirus infections (including some with Sabin PV2) are being missed by the RD-L20B-cell-culture-based algorithm, thus highlighting the value of the RT-snPCR assay and its modifications. The circulation and preponderance of EV-C in Nigeria was also confirmed.






Similar content being viewed by others
References
Oberste MS, Maher K, Kilpatrick DR, Pallansch MA (1999) Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73(3):1941–1948
Oberste MS, Maher K, Flemister MR, Marchetti G, Kilpatrick DR, Pallansch MA (2000) Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol 38(3):1170–1174
World Health Organisation (1988) Global eradication of poliomyelitis by the year 2000 (World Health Assembly resolutionWHA41–28). http://www.polioeradication.org/content/publications/19880513_resolution.pdf
Nathanson N, Kew OM (2010) From emergence to eradication: the epidemiology of poliomyelitis deconstructed. Am J Epidemiol 172(11):1213–1229
World Health Organisation (2003) Guidelines for environmental surveillance of poliovirus circulation. World Health Organisation, Geneva
World Health Organisation (2004) Polio laboratory Manual, 4th edn. World Health Organisation, Geneva
Sadeuh-Mba SA, Bessaud M, Massenet D, Joffret ML, Endegue MC, Njouom R, Reynes JM, Rousset D, Delpeyroux F (2013) High frequency and diversity of species C enteroviruses in Cameroon and neighboring countries. J Clin Microbiol 51:759–770
Adeniji JA, Faleye TOC (2014) Impact of cell lines included in enterovirus isolation protocol on perception of nonpolio enterovirus species C diversity. J Virol Methods. doi:10.1016/j.jviromet.2014.07.016
Faleye TOC, Adeniji JA (2015) Nonpolio enterovirus-C (NPEV-C) strains circulating in South-Western Nigeria and their contribution to the emergence of recombinant cVDPV2 lineages. Brit J Virol 2(5):68–73
Nix WA, Oberste MS, Pallansch MA (2006) Sensitive, seminested PCR amplification of VP1 sequeances for direct Identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 44(8):2698–2704
World Health Organisation (2015) Enterovirus surveillance guidelines: guidelines for enterovirus surveillance in support of the Polio Eradication Initiative. World Health Organisation, Geneva
Rahimi P, Tabatabaie H, Gouya MM, Mahmudi M, Musavi T, Rad KS, Azad TM, Nategh R (2009) Direct identification of non-polio enteroviruses in residual paralysis cases by analysis of VP1 sequences. J Clin Virol 45(2):139–141
Sadeuh-Mba SA, Bessaud M, Joffret ML, Endegue Zanga MC, Balanant J, MpoudiNgole E, Njouom R, Reynes JM, Delpeyroux F, Rousset D (2014) Characterization of enteroviruses from non-human primates in cameroon revealed virus types widespread in humans along with candidate new types and species. PLoS Negl Trop Dis 8(7):e3052. doi:10.1371/journal.pntd.0003052
Faleye TOC, Adewumi MO, Coker BA, Nudamajo FY, Adeniji JA (2016) Direct detection and identification of enteroviruses from faeces of healthy Nigerian children using a cell-culture independent RT-seminested PCR assay. Adv Virol. doi:10.1155/2016/1412838
Faleye TOC, Adewumi MO, Kareem SA, Adesuyan YO, Fapohunda FA, Fasanya ST, Jimeto T, Lawrence OE, Obembe AA, Adeniji JA (2016) The impact of a panenterovirus VP1 assay on our perception of the enterovirus diversity landscape of a sample. J Hum Virol. Retrovirol 4(3):00134
Kroneman A, Vennema H, Deforche K, Van der Avoort H, Penarandac S, Oberste MS, Vinjéc J, Koopmans M (2011) An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol 51:121–125
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Adeniji JA, Faleye TOC (2015) Enterovirus C strains circulating in Nigeria and their contribution to the emergence of recombinant circulating vaccine-derived polioviruses. Adv Virol 160(3):675–683
Jegouic S, Joffret ML, Blanchard C, Riquet FB, Perret C, Pelletier I, Colbere-Garapin F, Rakoto-Andrianarivelo M, Delpeyroux F (2009) Recombination between polioviruses and co-circulating Coxsackie A viruses: role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathog 5(5):E1000412
Bessaud M, Joffret ML, Holmblat B, Razafindratsimandresy R, Delpeyroux F (2011) Genetic relationship between cocirculating human enteroviruses species C. PLoS One 6(9):E24823
Di Cristianzano V, Boettcher S, Diedrich S, Timmen-Wego M, Knops E, Luebke N, Kaiser R, Pfister H, Kabore Y, D’Alfonso R (2015) Detection and characterization of enteroviruses and parechoviruses in healthy people living in the South of Cote d’Ivoire. J Clin Virol 71:40–43
Smura T, Blomqvist S, Vuorinen T, Ivanova O, Samoilovich E, Al-Hello H, Savolainen-Kopra C, Hovi T, Roivainen M (2014) The evolution of Vp1 gene in enterovirus C species sub-group that contains types CVA-21, CVA-24, EV-C95, EV-C96 and EV-C99. PLoS One 9(4):E93737
Adeniji JA, Faleye TOC (2014) Isolation and identification of enteroviruses from sewage and sewage contaminated water in Lagos, Nigeria. Food Environ Virol 6:75–86
Oyero OG, Adu FD, Ayukekbong JA (2014) Molecular characterization of diverse species enterovirus-B types from children with acute flaccid paralysis and asymptomatic children in Nigeria. Virus Res 189:189–193
Burns CC, Diop OM, Sutter RW, Kew OM (2014) Vaccine derived polioviruses. JID 210(suppl 1):S283–S293
World Health Organisation (2014) WHO global action plan to minimize poliovirus facility-associated risk, 3rd edn. World Health Organisation, Geneva
Adeniji JA, Oragwa AO, George UE, Ibok UI, Faleye TOC, Adewumi MO (2016) Enterovirus A119 in a child with acute flaccid paralysis. Nigeria. J Hum Virol Retrovirol 4(2):00126
Zhang Y, Hong M, Sun Q, Zhu S, Li X, Yan D, Wang D, Xu W, Li X (2014) Molecular typing and characterization of a new serotype of human enterovirus (EV-B111) identified in China. Virus Res 183(2014):75–80
Oberste MS, Feeroz MM, Maher K, Nix WA, Engel GA, Hasan KM, Begum S, Oh G, Chowdhury AH et al (2013) Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J Virol 87:558–571
Acknowledgements
We thank the WHO National Polio Laboratory in Ibadan, Nigeria, for providing the anonymous samples analyzed in this study.
Author information
Authors and Affiliations
Contributions
1. Study design (JAA, MOA, TOCF). 2. Sample collection, laboratory and data analysis (all authors). 3. Wrote, revised, read and approved the final draft of the manuscript (all authors).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists.
Ethical approval
There was no contact with human participants by any of the authors, and the article does not contain any information that can be used to associate the enterovirus types analyzed in this study to any individual.
Funding
This study was funded by contributions from the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3466_MOESM1_ESM.docx
Supplementary Table 1: Estimation of sensitivity of the RD and L20B cell lines used in the WHO-accredited polio laboratory (polio lab) in Ibadan, Nigeria. These sensitivity data were generated following the method described in the guidelines for poliovirus surveillance (WHO, 2004) and is for the period (August 2015) during which the negative samples analyzed in this study were screened by the polio lab. Hence, these data were not generated by the authors of this study but were generously provided on retrospective request from the archives of the polio lab. (DOCX 11 kb)
Rights and permissions
About this article
Cite this article
Adeniji, J.A., Oragwa, A.O., George, U.E. et al. Preponderance of enterovirus C in RD-L20B-cell-culture-negative stool samples from children diagnosed with acute flaccid paralysis in Nigeria. Arch Virol 162, 3089–3101 (2017). https://doi.org/10.1007/s00705-017-3466-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3466-2