Abstract
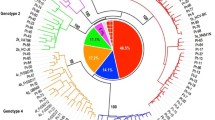
Next-generation sequencing (NGS) provides a practical approach to HCV complete-genome sequencing, detecting low-frequency variants and allowing analysis of viral genetic diversity (quasispecies) in the sample, and so far, it is very useful for identifying preexisting drug-resistant mutants and emerging escape mutations, as well as detecting viral recombinants containing genomic regions from different genotypes and subtypes. The aim of this study was to analyze the complete coding region of hepatitis C virus (HCV) genotype 1 (subtypes 1a and 1b) from patients with chronic infection who were direct-acting antiviral (DAA) naïve. Next-generation sequencing (Ion Torrent™ PGM) was used to determine the sequence of the complete coding region of 100 HCV-monoinfected DAA-naïve patients (51 and 49 subtypes 1a and 1b, respectively). We report the first description of nearly complete HCV genome sequences of subtype 1a and 1b isolates from a large population of Brazilian patients with chronic hepatitis C, and HCV-1a grouped in two different clades. Using this methodology, an inter-subtype 1a/1b recombinant was identified in this study.




Similar content being viewed by others
References
Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST (2013) Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342
Timm J, Roggendorf M (2007) Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J Gastroenterol 13:4808–4817
Alavian SM, Ande SR, Coombs KM, Yeganeh B, Davoodpour P, Hashemi M, Los M, Ghavami S (2011) Virus-triggered autophagy in viral hepatitis—possible novel strategies for drug development. J Viral Hepat 18:821–830
Lee LY, Tong CY, Wong T, Wilkinson M (2012) New therapies for chronic hepatitis C infection: a systematic review of evidence from clinical trials. Int J Clin Pract 66:342–355
Yang PL, Gao M, Lin K, Liu Q, Villareal VA (2011) Anti-HCV drugs in the pipeline. Curr Opin Virol 1:607–616
Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P (2014) Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327
Luciani F, Bull RA, Lloyd AR (2012) Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol 30:443–452
Plauzolles A, Lucas M, Gaudieri S (2013) Hepatitis C virus adaptation to T-cell immune pressure. Sci World J 2013:673240
Xu CH, Shen T, Zheng JJ, Tu J, Zhang WD, Lu FM (2012) Higher dN/dS ratios in the HCV core gene, but not in the E1/HVR1 gene, are associated with human immunodeficiency virus-associated immunosuppression. Arch Virol 157:2153–2162
Morel V, Ghoubra F, Izquierdo L, Martin E, Oliveira C, Francois C, Brochot E, Helle F, Duverlie G, Castelain S (2016) Phylogenetic analysis of a circulating hepatitis C virus recombinant strain 1b/1a isolated in a French hospital centre. Infect Genet Evol 40:374–380
Kalinina O, Norder H, Mukomolov S, Magnius LO (2002) A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol 76:4034–4043
Kageyama S, Agdamag DM, Alesna ET, Leano PS, Heredia AM, Abellanosa-Tac-An IP, Jereza LD, Tanimoto T, Yamamura J, Ichimura H (2006) A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J Med Virol 78:1423–1428
Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus AD, Haagmans BL (2006) Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol 80:7569–7577
Legrand-Abravanel F, Claudinon J, Nicot F, Dubois M, Chapuy-Regaud S, Sandres-Saune K, Pasquier C, Izopet J (2007) New natural intergenotypic (2/5) recombinant of hepatitis C virus. J Virol 81:4357–4362
Lee YM, Lin HJ, Chen YJ, Lee CM, Wang SF, Chang KY, Chen TL, Liu HF, Chen YM (2010) Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J Med Virol 82:57–68
Colina R, Casane D, Vasquez S, Garcia-Aguirre L, Chunga A, Romero H, Khan B, Cristina J (2004) Evidence of intratypic recombination in natural populations of hepatitis C virus. J Gen Virol 85:31–37
Ross RS, Verbeeck J, Viazov S, Lemey P, Van Ranst M, Roggendorf M (2008) Evidence for a complex mosaic genome pattern in a full-length hepatitis C virus sequence. Evol Bioinform Online 4:249–254
Moreno P, Alvarez M, Lopez L, Moratorio G, Casane D, Castells M, Castro S, Cristina J, Colina R (2009) Evidence of recombination in hepatitis C virus populations infecting a hemophiliac patient. Virol J 6:203
Calado RA, Rocha MR, Parreira R, Piedade J, Venenno T, Esteves A (2011) Hepatitis C virus subtypes circulating among intravenous drug users in Lisbon, Portugal. J Med Virol 83:608–615
Garson JA, Ring C, Tuke P, Tedder RS (1990) Enhanced detection by PCR of hepatitis C virus RNA. Lancet 336:878–879
Alcantara LC, Cassol S, Libin P, Deforche K, Pybus OG, Van Ranst M, Galvao-Castro B, Vandamme AM, de Oliveira T (2009) A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 37:W634–W642
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Cuypers L, Snoeck J, Vrancken B, Kerremans L, Vuagniaux G, Verbeeck J, Nevens F, Camacho RJ, Vandamme AM, Van Dooren S (2014) A near-full length genotypic assay for HCV1b. J Virol Methods 209:126–135
Capobianchi MR, Giombini E, Rozera G (2013) Next-generation sequencing technology in clinical virology. Clin Microbiol Infect 19:15–22
Peres-da-Silva A, Almeida AJ, Lampe E (2012) Genetic diversity of NS3 protease from Brazilian HCV isolates and possible implications for therapy with direct-acting antiviral drugs. Mem Inst Oswaldo Cruz 107:254–261
Lampe E, Lewis-Ximenez L, Espirito-Santo MP, Delvaux NM, Pereira SA, Peres-da-Silva A, Martins RM, Soares MA, Santos AF, Vidal LL, Germano FN, de Martinez AM, Basso R, Pinho JR, Malta FM, Gomes-Gouvea M, Moliterno RA, Bertolini DA, Fujishima MA, Bello G (2013) Genetic diversity of HCV in Brazil. Antivir Ther 18:435–444
Wyles DL, Gutierrez JA (2014) Importance of HCV genotype 1 subtypes for drug resistance and response to therapy. J Viral Hepat 21:229–240
Shi W, Freitas IT, Zhu C, Zheng W, Hall WW, Higgins DG (2012) Recombination in hepatitis C virus: identification of four novel naturally occurring inter-subtype recombinants. PLoS One 7:e41997
Gonzalez-Candelas F, Lopez-Labrador FX, Bracho MA (2011) Recombination in hepatitis C virus. Viruses 3:2006–2024
Yun Z, Lara C, Johansson B, Lorenzana de Rivera I, Sonnerborg A (1996) Discrepancy of hepatitis C virus genotypes as determined by phylogenetic analysis of partial NS5 and core sequences. J Med Virol 49:155–160
Du H, Qi Y, Hao F, Huang Y, Mao L, Ji S, Huang M, Qin C, Yan R, Zhu X, Zhang C (2012) Complex patterns of HCV epidemic in Suzhou: evidence for dual infection and HCV recombination in East China. J Clin Virol 54:207–212
Hedskog C, Doehle B, Chodavarapu K, Gontcharova V, Crespo Garcia J, De Knegt R, Drenth JP, McHutchison JG, Brainard D, Stamm LM, Miller MD, Svarovskaia E, Mo H (2015) Characterization of hepatitis C virus intergenotypic recombinant strains and associated virological response to sofosbuvir/ribavirin. Hepatology 61:471–480
Lauck M, Alvarado-Mora MV, Becker EA, Bhattacharya D, Striker R, Hughes AL, Carrilho FJ, O’Connor DH, Pinho JR (2012) Analysis of hepatitis C virus intrahost diversity across the coding region by ultradeep pyrosequencing. J Virol 86:3952–3960
Miura M, Maekawa S, Takano S, Komatsu N, Tatsumi A, Asakawa Y, Shindo K, Amemiya F, Nakayama Y, Inoue T, Sakamoto M, Yamashita A, Moriishi K, Enomoto N (2013) Deep-sequencing analysis of the association between the quasispecies nature of the hepatitis C virus core region and disease progression. J Virol 87:12541–12551
Akuta N, Suzuki F, Fukushima T, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Hara T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H (2013) Prediction of treatment efficacy and telaprevir-resistant variants after triple therapy in patients infected with hepatitis C virus genotype 1. J Clin Microbiol 51:2862–2868
Svarovskaia ES, Martin R, McHutchison JG, Miller MD, Mo H (2012) Abundant drug-resistant NS3 mutants detected by deep sequencing in hepatitis C virus-infected patients undergoing NS3 protease inhibitor monotherapy. J Clin Microbiol 50:3267–3274
Prosperi MC, Yin L, Nolan DJ, Lowe AD, Goodenow MM, Salemi M (2013) Empirical validation of viral quasispecies assembly algorithms: state-of-the-art and challenges. Sci Rep 3:2837
Fan X, Di Bisceglie AM (2010) RT-PCR amplification and cloning of large viral sequences. Methods Mol Biol 630:139–149
Ninomiya M, Ueno Y, Funayama R, Nagashima T, Nishida Y, Kondo Y, Inoue J, Kakazu E, Kimura O, Nakayama K, Shimosegawa T (2011) Use of illumina deep sequencing technology to differentiate hepatitis C virus variants. J Clin Microbiol 50:857–866
Acknowledgments
Special thanks to the patients, to Dr Maria C. M. Correa for providing samples, and to Dr Ester C. Sabino for allowing the use of next-generation sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Financial support
This project was supported by grant 443152/2014-4 (CNPq) and Alves de Queiroz Family Fund for Research. Karine Vieira Gaspareto is a graduate student from Programa de Pós-Graduação em Biotecnologia, USP, and received a fellowship from CAPES. João Renato Rebello Pinho, Ester Cerdeira Sabino and Maria Cassia Mendes Correa received a fellowship from CNPq (Bolsista de Produtividade).
Rights and permissions
About this article
Cite this article
Gaspareto, K.V., Ribeiro, R.M., de Mello Malta, F. et al. HCV inter-subtype 1a/1b recombinant detected by complete-genome next-generation sequencing. Arch Virol 161, 2161–2168 (2016). https://doi.org/10.1007/s00705-016-2889-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2889-5