Abstract
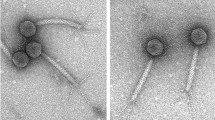
Bacillus cereus group-specific bacteriophage BCP8-2 exhibits a broad lysis spectrum among food and human isolates (330/364) of B. cereus while not infecting B. subtilis (50) or B. licheniformis (12) strains. Its genome is 159,071 bp long with 220 open reading frames, including genes for putative methyltransferases, metallo-beta-lactamase, and a sporulation-related SpoIIIE homolog, as wells as 18 tRNAs. Comparative genome analysis showed that BCP8-2 is related to the recently proposed Bastille-like phages, but not with either SPO1-like or Twort-like phages of the subfamily Spounavirinae.

Similar content being viewed by others
References
Bandara N, Jo J, Ryu S, Kim K-P (2012) Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol 31:9–16
Barylski J, Nowicki G, Goździcka-Józefiak A (2014) The discovery of phiAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae. PLoS ONE 9:e86632
Bebrone C (2007) Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 74:1686–1701
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
El-Arabi TF, Griffiths MW, She Y-M, Villegas A, Lingohr EJ, Kropinski AM (2013) Genome sequence and analysis of a broad-host range lytic bacteriophage that infects the Bacillus cereus group. Virol J 10:48
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Lee J-H, Shin H, Ryu S (2014) Characterization and comparative genomic analysis of bacteriophages infecting members of the Bacillus cereus group. Arch Virol 159:871–884
Mahadevan P, King JF, Seto D (2009) CGUG: in silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res Notes 2:168
Nakamura Y, Gojobori T, Ikemura T (2000) Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res 28:292
Omasits U, Ahrens CH, Müller S, Wollscheid B (2013) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886
Pope WH, Anders KR, Baird M et al (2014) Cluster M mycobacteriophages Bongo, PegLeg, and Rey with unusually large repertoires of tRNA isotypes. J Virol 88:2461–2480
Ryazanova AY, Abrosimova L, Oretskaya T, Kubareva E (2012) Diverse domains of (cytosine-5)-DNA methyltransferases: structural and functional characterization. In: Dricu A (ed) Methylation—from DNA, RNA and histones to disease and treatment. InTech, Croatia, pp 29–69
Santos SB, Kropinski AM, Ceyssens P-J, Ackermann H-W, Villegas A, Lavigne R, Krylov VN, Carvalho CM, Ferreira EC, Azeredo J (2011) Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J Virol 85:11265–11273
Schuch R, Pelzek AJ, Fazzini MM, Nelson DC, Fischetti VA (2014) Complete genome sequence of Bacillus cereus sensu lato bacteriophage Bcp1. Genome Announc 2:e00334-00314
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010
Yee LM, Matsumoto T, Yano K, Matsuoka S, Sadaie Y, Yoshikawa H, Asai K (2011) The genome of Bacillus subtilis phage SP10: a comparative analysis with phage. Biosci Biotechnol Biochem 75:944–995
Acknowledgments
This work was supported by the High Value-Added Food Technology Development Program (Project No. 112108-2 and 313037-3), Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) of Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
P. T. Asare and N. Bandara contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Asare, P.T., Bandara, N., Jeong, TY. et al. Complete genome sequence analysis and identification of putative metallo-beta-lactamase and SpoIIIE homologs in Bacillus cereus group phage BCP8-2, a new member of the proposed Bastille-like group. Arch Virol 160, 2647–2650 (2015). https://doi.org/10.1007/s00705-015-2548-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2548-2