Abstract
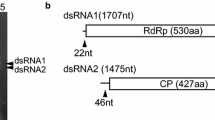
The bisegmented genome of a putative double-stranded (ds) RNA virus from Ustilaginoidea virens was sequenced and analyzed. The larger genomic segment of 2112 bp encodes a putative RNA-dependent RNA polymerase (RdRp, 628 aa), and the smaller one of 2082 bp encodes a putative coat protein (CP) of 539 aa. The 5′ untranslated regions (UTR) of the two segments share regions of high sequence homology. Phylogenetic analysis indicates that this novel partitivirus, named Ustilaginoidea virens partitivirus 2 (UvPV2), can be assigned to the family Partitiviridae.


Similar content being viewed by others
References
Tanaka E, Ashizawa T, Sonoda R, Tanaka C (2008) Villosiclava virens gen. nov., comb. nov., the teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon 106:491
White JF Jr, Sullivan R, Moy M, Patel R, Duncan R (2000) An overview of problems in the classification of plant-parasitic Clavicipitaceae. Stud Mycol 45:95–105
Bischoff J, Sullivan R, Kjer K, White J (2004) Phylogenetic placement of the anamorphic tribe Ustilaginoideae (Hypocreales, Ascomycota). Mycologia 96(5):1088–1094
Cooke M (1878) Some extra-European fungi. Grevillea 7:13–15
Osada S (1995) Effect of false smut occurrence on yield and quality of rice. Annual Report of the Society of Plant Protection of North Japan
Chiba S, Salaipeth L, Lin Y-H, Sasaki A, Kanematsu S, Suzuki N (2009) A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J Virol 83(24):12801–12812
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Ann Rev Phytopathol 47:353–384
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10(1):115–128
Mertens P (2004) The dsRNA viruses. Virus Res 101(1):3–13
Zhang T, Jiang Y, Huang J, Dong W (2013) Complete genome sequence of a putative novel victorivirus from Ustilaginoidea virens. Arch Virol 158(6):1403–1406
Zhang T, Jiang Y, Huang J, Dong W (2013) Genomic organization of a novel partitivirus from the phytopathogenic fungus Ustilaginoidea virens. Arch Virol 158(11):2415–2419
Dodds JA, Bar-Joseph M (1983) Double-stranded RNA from plants infected with closteroviruses. Phytopathology 73(3):419–423
Hunst PL, Latterell FM, Rossi AE (1986) Variation in double-stranded RNA from isolates of Pyricularia oryzae. Phytopathology 76(7):674–678
Froussard P (1993) rPCR: a powerful tool for random amplification of whole RNA sequences. Genome Research 2(3):185–190
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90(6):1423–1432
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Strauss EE, Lakshman DK, Tavantzis SM (2000) Molecular characterization of the genome of a partitivirus from the basidiomycete Rhizoctonia solani. J Gen Virol 81(2):549–555
Yaegashi H, Nakamura H, Sawahata T, Sasaki A, Iwanami Y, Ito T, Kanematsu S (2013) Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, Rosellinia necatrix, in an apple orchard. FEMS Microbiol Ecol 83(1):49–62
Huang S, Ghabrial SA (1996) Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc Natl Acad Sci USA 93(22):12541–12546
Acknowledgments
This work was supported by the Special Fund for Agro-Scientific Research in the Public Interest (200903039-3) from the Ministry of Agriculture of China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplementary Fig. S1 Phylograms of the UvPV2 CP. Multiple sequence alignments of the deduced amino acid sequences of UvPV2 and selected viruses belonging to families Chrysoviridae, Totiviridae and Partitiviridae were performed using MEGA 5.0. The phylogenic trees were analyzed using the NJ method with 1000 bootstrap replicates. The abbreviations of other virus names and GenBank accession numbers are as follows: Amasya cherry disease associated chrysovirus (ACDACV; YP-001531162); Aspergillus fumigatus chrysovirus (AfuCV; CAX48751); Botryotinia fuckeliana partitivirus 1 (BfPV1;YP-001109579); cherry chlorotic rusty spot associated chrysovirus (CCRSACV; AH03665); Coniothyrium minitans mycovirus (CmRV; YP-392466); Epichloe festucae virus 1 (EfV1; CAK02787); Heterobasidion RNA virus 8 (HetRV8; AFW17811); Helminthosporium victoriae 145S (HV145SV; YP-052859); Helicobasidium mompa totivirus 1-17 (HmT1-17; NC-005074); red clover cryptic virus 2 (RCCV-2; YP_007889824); Sphaeropsis sapinea RNA virus 2 (SsRV2; NP-047559); pepper cryptic virus 2 (PCV-2; AEJ07893); Primula malacoides virus China/Mar2007 (PmV1; EU195327).
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, Y., Luo, C., Jiang, D. et al. The complete genomic sequence of a second novel partitivirus infecting Ustilaginoidea virens . Arch Virol 159, 1865–1868 (2014). https://doi.org/10.1007/s00705-014-1991-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-014-1991-9