Abstract
To date, most members of the Siphoviridae family of bacteriophages remain unclassified, including the 46 staphylococcal phages for which the complete genome sequences have been deposited in public databases. Comparative nucleotide and protein sequence analysis, in addition to available data on phage morphology, allowed us to propose three new phage genera within the family Siphoviridae: “3alikevirus”, “77likevirus” and “Phietalikevirus”, which include related phages infecting Staphylococcus aureus and Staphylococcus epidermidis. However, six phages infecting S. aureus, Staphylococcus pasteuri, Staphylococcus hominis and Staphylococcus capitis strains remain to be classified (orphan phages). Overall, the former phages share morphological features and genome organization. The three groups have conserved domains containing peptidoglycan hydrolytic activities clearly identified as part of tape measure proteins (“3alikevirus” and “77likevirus”) or as individual virion-associated proteins (“Phietalikevirus”). In addition, bacteriophages belonging to the genus “3alikevirus” share closely related DNA-processing and packaging proteins, while bacteriophages included in the genus “Phietalikevirus” encode specific tail proteins for host interaction. These properties are considered distinctive for these genera. Orphan phages seem to have a more divergent organization, but they share some properties with members of these proposed genera.
Similar content being viewed by others
Introduction
The recent increase in phage genome sequences available in public databases (828 in the order Caudovirales as of June 20, 2013) along with the development of bioinformatic tools has enabled the classification of bacteriophages on the basis of their genetic relationships. Recent approaches led to the creation of the subfamilies Autographivirinae and Picovirinae in the family Podoviridae and the subfamily Tevenvirinae within the family Myoviridae [1–3]. To date, a large number of Staphylococcus aureus phages have been described, but a complete genetic and functional characterization has been performed only on a limited number. Previous attempts to classify S. aureus phages were based on genome sizes and resulted in three classes [4]. Class I included phages with a genome size <20 kb (P68, 66, and 44AHJD); class II, 20-40 kb (42e, 3A, 47, 187, 69, 53, 85, 2638A, 77, 37, 96, 71, 55, 29, 52A, 88, 92, EW and X2); and class III, 40-125 kb (Twort, K, and G1). Morphological differences among these classes were also observed. Class I phages belong to the family Podoviridae (C1 morphotype) with a short, non-contractile tail and an isometric head, whereas class II phages belong to the family Siphoviridae. The majority of class II phages show a B1 morphotype with a long, non-contractile tail and an isometric head. In this group, phages 42e, 3A, and 47 display a B2 morphotype with a long, non-contractile tail and elongated head. Finally, the three phages from class III are structurally defined as members of the family Myoviridae.
Recently, three novel temperate Siphoviridae phages (StB12, StB27, and StB20) from the coagulase-negative species (CoNS) Staphylococcus hominis and Staphylococcus capitis were isolated and characterized [5]. Based on the comparative analysis of these phages with prophage sequences, the authors proposed a novel classification for staphylococcal phages in nine different clusters.
The aim of the current work is to find a rational scheme for staphylococcal phage classification, which condenses the extensive data derived from genetic and proteomic studies to give some clues about the evolutionary relationships among these phages. Thus, we propose the creation of three new siphovirus genera named “3alikevirus”, “77likevirus” and “Phietalikevirus”, in which 46 staphylococcal phages have been grouped.
Comparative genomics
Bioinformatic analysis of 46 staphylococcal phages indicated relatively minor differences in genome sizes and G+C content, which range between 40-45.8 kb and 33-38 %, respectively. We first turned to comparative analysis employing BLASTN (http://blast.ncbi.nlm.nih.gov/); EMBOSS Stretcher (http://www.ebi.ac.uk/Tools/psa/emboss_stretcher/) and progressive-Mauve (http://gel.ahabs.wisc.edu/mauve/), along with protein content comparisons using CoreGenes 3.0 [6–8]. Before the global alignments could be performed, the genomes were manually colinearized, placing the arbitrary starting point for the sequence 150 bp in front of the gene for the small terminase subunit. The progressiveMauve alignments reveal a similar organization of the phages in modules that fit to the general structure of most double-stranded DNA bacteriophages [9]. These modules include packaging, structure/morphogenesis, host lysis, lysogeny and replication/regulation (Supplementary Figures 1-5). Taking into account phage morphology, genome size and organization, gene synteny, and a shared protein content of 40 %, three groupings can be described, for which we propose the status of genus, consistent with other Caudovirales groupings (Table 1): “3alikevirus” (3A, Φ12, 47, phiIPLA35, ΦSLT and 42e), “77likevirus” (77, ΦPVL108 and Φ13), and “Phietalikevirus” (ΦETA, 52A, 80, 29, 71, 55, ΦMR11, ΦETA3, 88, 92, X2, ΦNM4, 96, ΦETA2, Φ11, 53, 69, 80α, 85, ΦMR25, ΦNM1, ΦNM2, phiIPLA88, 187, SAP-26). Phages 2638A, TEM123, SpaA1, StB12, StB20, and StB27 did not reveal a clear relationship to the others and remain as orphan phages. Using the Needleman-Wunsch rapid global alignment algorithm of EMBOSS Stretcher, nucleotide similarities with the type phage in each genus have been computed. Values over 53 % for DNA sequence identity were observed within each proposed genus (Table 1). Between genera these values were lower than 50 % (data not shown). Focusing on the percentage of shared proteins with phage type, values over 42 % were observed within each genus, while lower values were determined between genera (data not shown).
Features of the bacteriophages belonging to the proposed genus “3alikevirus”
The proposed genus “3alikevirus” includes six of the staphylococcal phages. All of these viruses were reported to possess a prolate capsid (siphovirus B2 morphotype) that is 100 nm long and 50 nm wide, and a tail of 300-400 nm (for references, see Table 1). Comparative proteomic analysis using CoreGenes 3.0 and BLASTP indicates the presence of several group-specific proteins. Among the replication-associated proteins, there are a helicase-like protein and an A-type DNA polymerase. In the morphogenesis module, a unique capsid protein containing a pfam05065 motif (phage capsid family) and a major_cap_HK97 (TIGR01554; phage major capsid protein, HK97 family) motif can be distinguished. The tail module contains two specific proteins; the first shows a Sipho_tail (pfam05709) motif, while the second corresponds to a predicted prophage endopeptidase tail motif (pfam06605). Another protein that is exclusive for these phages is RinA, whereas the RinB homolog can be found in other phages.
Comparative analysis of DNA sequences using progressiveMauve alignments also indicates a particularly high degree of conservation in a region spanning the DNA-packaging proteins (small and large terminase subunits, portal and scaffolding proteins) (Supplementary Figure 1). In addition, all phages belonging to the genus “3alikevirus”, share a region encoding a putative nuclease (HNH superfamily).
A relevant characteristic of the genus “3alikevirus” is the presence of peptidoglycan hydrolytic domains included in their tape measure proteins (TMPs). Specifically, a lytic transglycosylase SLT domain and a peptidase_M23 domain are detected in all of them (Table 2).
Features of the bacteriophages belonging to the proposed genus “77likevirus”
The proposed genus “77likevirus” includes three S. aureus phages with a similar genome size and morphology (B1 morphotype). No data on the size of the viral particles have been published. These phages share characteristics with the other genera (“3alikevirus” and “Phietalikevirus”), such as the presence of nucleases in phages 77 and Φ13, similar to those from the “3alikevirus” genus, and a common morphotype with “Phietalikevirus”. Among the peptidoglycan hydrolase domains, a lytic transglycosylase SLT domain and a peptidase_M23 domain, similar to those found in “3alikevirus”, were detected in the tail tape measure proteins (TMPs) of the three phages belonging to the genus “77likevirus” (Table 2) (Supplementary Figure 2). No other structural proteins with peptidoglycan catalytic domains were detected in these phages.
Features of the bacteriophages belonging to the proposed genus “Phietalikevirus”
The proposed genus “Phietalikevirus”, which includes 31 staphylococcal phages, can theoretically be divided into three subgroups (Supplementary Figures 3-5) based on DNA similarity. The genome size of these phages is very similar among all of them. Virion morphology is nearly identical, with isometric heads of about 50 nm in diameter and a tail 175 nm long. All phages belonging to this genus show a B1 morphotype, with the exception of phage EW, which was described as having an A morphotype [4]. Due to its high homology with B1 bacteriophages, we conclude that this is incorrect.
A particularly interesting region of the “Phietalikevirus” phages is the sequence of genes in the tail morphogenesis module (Fig. 1). Five arrangements with small differences were observed. In most of the phages from subgroup 1 and subgroup 2 (ΦETA, 52A, 80, 29, 71, 55, ΦMR11, ΦETA3, 88, 92, X2, ΦNM4, 96, ΦETA2, Φ11, 53, 69, 80α, 85, ΦMR25, ΦNM1, ΦNM2, and phiIPLA88), this region is composed of a protein belonging to the SGNH hydrolase superfamily, a hypothetical protein, a tail protein, a virion-associated peptidoglycan hydrolase and a tail fiber protein (Fig. 1A). For most phages belonging to subgroup 3 (37, CNPH82, PH15, phiIPLA5 and phiIPLA7) a pectin-lyase-encoding gene can be observed downstream of the SGNH protein (Fig. 1B and C). In all these phages, with the exception of phages phiIPLA7 and PH15, a tail protein is also present. Notable differences can be seen in the makeup of this region for phages 187 and EW, which lacks the pectin lyase. No tail protein could be identified on phage 187 (Fig. 1D and E). The SGNH hydrolases containing lipase and esterase domains are present in all phages within the genus, including phage 187, which has a truncated protein (Fig. 1D). Moreover, peptidoglycan hydrolytic activities are present in all phages as virion-associated proteins containing two catalytic domains: a CHAP domain and a glucosaminidase domain (Fig. 1; Table 2). All phages except phiIPLA7, PH15 and phage 187 encode tail fiber proteins with a collagen helix domain downstream from the peptidoglycan hydrolases. This domain consists of a triple helix formed by repetitions of the amino acid sequence glycine-X-Y. A shorter sequence of this protein is also observed for phages phiIPLA5 and CNPH82. The members of subgroup 3, with the exception of phage 187 (Fig. 1B, C and D), have a pre-neck appendage protein between the SGNH hydrolase and the tail protein endowed with a pectin lyase and a peptidase domain.
Morphogenetic tail region in members of the proposed genus “Phietalikevirus”. (A) Structure for phages ΦETA, 80, 52A, 29, 55, ΦMR11, ΦETA3, ΦNM4, 96, Φ11, ΦNM1, ΦNM2, 69, 80α, 53, ΦETA2, ΦMR25, 85, phiIPLA88, 92, X2, 88, 71. (B) Structure shared by phiIPLA5, CNPH82 and phage 37. (C) Structure for phiIPLA7 and PH15. (D) and (E) Structure for phage 187 and EW, respectively. A dashed arrow represents a region with a variable number (from 3 to 5) of conserved hypothetical proteins
Orphan phages infecting Staphylococcus within the family Siphoviridae
Six siphophages infecting Staphylococcus have no clear homology to members of the previously proposed genera. For instance, regarding the peptidoglycan hydrolytic activities specific to the other genera, the orphan phages, with the exception of 2638A and StB20, lack lytic domains associated with the TMPs. Phage 2638A has a TMP containing only the peptidase_M23 domain but not a transglycosylase SLT domain. An interesting structure can be observed in S. capitis phage StB20, which encodes TMP containing the peptidoglycan hydrolytic domains (a peptidase_M23 domain and a transglycosylase SLT domain) and additionally, a truncated virion-associated peptidoglycan hydrolase bearing a CHAP domain (Table 2). Complete peptidoglycan hydrolases are present in phages TEM123 and StB27. Other phages, like StB12, encode potentially truncated PG hydrolases with a glucosaminidase domain. In phage SpaA1, no peptidoglycan lytic activities could be detected, either in the TMP or any other structural proteins.
Bacteriophage TEM123 shares characteristics with members of the proposed genus “Phietalikevirus”, such as the presence of structural proteins related to host interaction (SGNH and tail fiber protein) but lacks overall homology to these viruses (Table 2). Phage 2638A seems to be more related to the members of “3alikevirus”, sharing nuclease and helicase proteins (data not shown). Phages StB20 and StB27 may have a putative relationship to “Phietalikevirus” due to the presence of host-interaction proteins; StB27 has SGNH hydrolase and tail fiber protein, and StB20 also shows a tail fiber protein in the structural module (Table 2).
Phylogeny
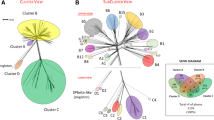
Typing of microbial pathogens is often performed using multi-locus sequence typing, in which fragments of several housekeeping genes are sequenced and concatenated and phylogenetic trees are constructed [10]. In this regard, using ClustalX [11], we have aligned the complete, colinearized genomes of all phages and generated a phylogenetic tree that was visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Figure 2 shows the whole-genome tree of the staphylococcal phages, which clearly reveals the three proposed genera, “3alikevirus”, “77likevirus” and “Phietalikevirus”, and the three subgroups belonging to the last of these genera being also recognizable. This tree also shows that phage TEM123 is related to the phietalikeviruses and shares over 40 % proteins with members of subgroup 2 of the genus (data not shown), but in spite of its shared protein content with ΦETA, their homology is only 28.8 % as calculated with CoreGenes. For these particular phages, the results of whole-genome phylogenetic tree building corroborate those of comparative genomic analysis, and this method can be a good tool in phage taxonomy.
Phylogenetic analysis of some conserved proteins was also carried out. Homology between phage large terminase subunits was previously shown to be indicative of different functional classes [12]. We determined that there is a high degree of sequence identity between large terminase subunits in the genus “3alikevirus”. This protein possesses the typical domains from the terminase 1 superfamily, which are shared by 2638A, StB20, and phages belonging to “77likevirus” (Fig. 3A). Phages belonging to “Phietalikevirus” (with the exception of phage SAP-26) and the orphan phages SpaA1, StB12, StB27 and TEM123 show large terminase subunits associated with the terminase 3 superfamily. These data indicate a different packaging mechanism in phages of the different genera, which would further support the proposed clustering.
This grouping is further supported by phylogenetic analysis of the major head proteins. In fact, it shows a clear correlation between the proposed staphylococcal genera and even a higher homology to those belonging to the subgroups of the genus “Phietalikevirus” (Fig. 3B). Similar results were observed using head and tail proteins, such as portal protein and TMP (data not shown).
Discussion
The current phage taxonomy, as laid out by the International Committee on Taxonomy of Viruses (ICTV), is based on a multitude of phenotypic and genotypic parameters, including morphology, infectivity and genome organization. Several proposals for phage classification based on sequence information have been made, sometimes with conflicting views. Rohwer and Edwards [13] built a tree based on pairwise dissimilarities between the complete bacteriophage proteomes, Pride and colleagues [14] proposed a phylogeny based on tetranucleotide usage patterns, and Lima-Mendez and colleagues [15] published a framework for a reticulate classification of phages based on gene content, which incorporates more elements of mosaicisms and horizontal exchange [16–18]. More disruptive is the non-hierarchical model proposed by Lawrence and colleagues [19], which is not based in viral lineages but instead on the creation of reticulate groups that more accurately represent biological relationships among these phages. Phages could belong to several groups, and phages within a group would share a particular module or phenotypic characteristics, reflecting their mosaic nature. In this work, we confirm the validity of the CoreGenes approach to create coherent phage groups as well as the identification of individual phages with special properties. Thus, a detailed analysis of staphylococcal phages within the family Siphoviridae allowed us to propose three genera based on close genetic similarity and on the presence of specific proteins involved in DNA packaging and host interaction. In addition to the high sequence identity across the structural module [20], we have also observed gene synteny in the region comprising the DNA packaging proteins of phages grouped as “3alikevirus”. This feature was previously described for phages 3A, 47 and 42e by Kwan and colleagues [4]. Furthermore, phylogenetic analysis of both the terminase large subunit and the major head protein clearly substantiate and support the establishment of the genus “3alikevirus”. In addition, the predicted DNA packaging mechanism used by members of “3alikevirus” is mediated by cohesive ends and a specific large terminase subunit (terminase 1), and it differs from that of the members of “Phietalikevirus”. Phages belonging to the genera “3alikevirus” and “77likevirus” are likely to use a cos packaging mechanism, as has been suggested previously for phage phiIPLA35 and phage ΦSLT [21, 22]. By contrast, phietalikeviruses appear to use a pac site to package DNA into capsids, as has suggested for some phages such as phiIPLA88 and Φ11 [21, 23]. Clearly, the homology among large terminase subunits with similar enzymatic end-generating functions supported the proposed staphylococcal phage genera.
The presence of HNH superfamily (homing) nucleases seems to be specific to the “3alikevirus”. It has been proposed that the presence of endonuclease genes in phage genomes can be considered analogous to insertion of transposon elements in bacterial genomes [24, 25], which could explain their presence in phages infecting members of genera other than Staphylococcus as well as in orphan phages (StB20 and 2638A) and some 77likeviruses (data not shown). Moreover, highly conserved helicase proteins (DEAD-like helicases superfamily) found in members of “3alikevirus” are also present in phage 2638A.
Close examination of the differences between phage genera reveals two distinct arrangements in the peptidoglycan hydrolytic domains. Genomes of bacteriophages belonging to the genera “3alikevirus” and “77likevirus” encode peptidoglycan hydrolytic domains (peptidase_M23 and transglycosylase (SLT), as part of TMPs), whereas in “Phietalikevirus”, individual genes encode CHAP and glucosaminidase domains located in virion-associated peptidoglycan hydrolases. Previous analyses have revealed that phage TMPs frequently contain a soluble lytic transglycosylase domain [13, 26], and lytic transglycosylases have been proposed to be involved in phage DNA entry during early stages of infection [27, 28]. Similar catalytic domains were previously described in TMPs from several mycobacteriophages, where they play an essential role in the infection of stationary-phase cells [29]. In other phages, such as phage T5, these catalytic domains are included in a TMP that also carries fusogenic activities [30].
Phages of the proposed genus “Phietalikevirus” have peptidoglycan hydrolytic activity that is also associated with the phage particle, but as an independent protein. Virion-associated peptidoglycan hydrolases facilitate the entry of phage DNA across the bacterial cell envelope during infection [31]. They are also responsible for “lysis from without,” a phenomenon caused by some phages when adsorbed onto the host cell at very high numbers [32, 33]. Activity of these proteins against peptidoglycan was confirmed for those encoded by phages phiIPLA88 and ΦMR11 [34, 35]. Although the peptidoglycan hydrolase activity of these proteins has been demonstrated, its function in the phage lytic cycle is not clear. Likewise, PRD1 and T7 mutant phages without virion-associated peptidoglycan hydrolase activity are able to infect host cells, although the process is significantly delayed [31, 36].
The “Phietalikevirus” genus division is also supported by a more complex host interaction machinery, including a SGNH protein, a pectin lyase, a tail protein and a tail fiber protein. These proteins are probably involved in degradation of extracellular or capsular material, facilitating phage infection. SGNH proteins are involved in the hydrolysis of a wide variety of substrates, including fatty acids, aromatic esters, and amino acid derivatives [37]. Pectin lyase proteins have been described as putative hydrolases against extracellular components from bacterial biofilms [38]. They comprised two domains, including a pectin/lyase domain with a right-handed beta helical fold like those identified in carbohydrate depolymerizing enzymes [39]. These relationships suggest that these proteins might interact with the bacterial cell envelope and therefore might be related to bacterial receptor interaction.
It is interesting to highlight the structure of the orphan phages 2638A, StB12 and StB20. These phages are not clearly related to members of any of the proposed genera on the basis of DNA and protein homology. They have truncated peptidoglycan hydrolytic domains (2638A and StB12) or “duplicate” domains (StB20) that could be seen as intermediaries in exchange of these catalytic domains among phages.
Overall, our results are in accordance with previous attempts at classification of staphylococcal phages and prophages [4, 5]. The comparative nucleotide and protein sequence analyses performed have allowed us to define three novel genera in which phages grouped previously [4] have been included. Of note is the fact that the clusters proposed in the recent classification provided by Deghorain et al. [5] match, with a few exceptions, our classification in genera and subgroups, supporting its validity.
References
Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM (2008) Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res Microbiol 159:406–414
Lavigne R, Ceyssens PJ (2012) Order Caudovirales; family Myoviridae; genus “T4-like viruses”. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy—classification and nomenclature of viruses. Ninth report of the International Committee on taxonomy of viruses. Elsevier Academic Press, New York, pp 46–49
Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM (2009) Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 9:224. doi:10.1186/1471-2180-9-224
Kwan T, Liu J, DuBow M, Gros P, Pelletier J (2005) The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA 102:5174–5179
Deghorain M, Bobay LM, Smeesters PR, Bousbata S, Vermeersch M, Perez-Morga D, Drèze PA, Rocha EP, Touchon M, Van Melderen L (2012) Characterization of novel phages isolated in coagulase-negative staphylococci reveals evolutionary relationships with Staphylococcus aureus phages. J Bacteriol 194:5829–5839
Mahadevan P, King JF, Seto D (2009) CGUG: in silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res Notes 2:168
Mahadevan P, King JF, Seto D (2009) Data mining pathogen genomes using GeneOrder, CoreGenes and CGUG: gene order, synteny, and in silico proteomes. Int J Comput Biol Drug Des 2:100–114
Zafar N, Mazumder R, Seto D (2002) CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinforma 3:12
Brüssow H, Desiere F (2001) Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol 39:213–222
Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. PNAS 95:3140–3145
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW (2005) The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J Bacteriol 187:1091–1104
Rohwer F, Edwards R (2002) The phage proteomic tree: a genome-based taxonomy for phage. J Bacteriol 184:4529–4535
Pride DT, Wassenaar TM, Ghose C, Blaser MJ (2006) Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics 7:8
Lima-Mendez G, Van Helden J, Toussaint A, Leplae R (2008) Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol 25:762–777
Hendrix RW, Lawrence JG, Hatfull GF, Casjens S (2000) The origins and ongoing evolution of viruses. Trends Microbiol 8:504–508
Brüssow H, Hendrix RW (2002) Phage genomics: small is beautiful. Cell 108:13–16
Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF (1999) Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci USA 96:2192–2197
Lawrence JG, Hatfull GF, Hendrix RW (2002) Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol 184:4891–4905
Proux C, van Sinderen D, Suarez J, Garcia P, Ladero V, Fitzgerald GF, Desiere F, Brüssow H (2002) The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J Bacteriol 184:6026–6936
García P, Martínez B, Obeso JM, Lavigne R, Lurz R, Rodríguez A (2009) Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl Environ Microbiol 75:7663–7673
Narita S, Kaneko J, Chiba J, Piémont Y, Jarraud S, Etienne J, Kamio Y (2001) Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. Gene 268:195–206
Iandolo JJ, Worrell V, Groicher KH, Qian Y, Tian R, Kenton S, Dorman A, Ji H, Lin S, Loh P, Qi S, Zhu H, Roe BA (2002) Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene 289:109–118
Ceyssens PJ, Hertveldt H, Ackermann W, Noben JP, Demeke M, Volckaert G, Lavigne R (2008) The intron-containing genome of the lytic Pseudomonas phage LUZ24 resembles the temperate phage PaP3. Virology 377:233–238
Roberts MD, Martin NL, Kropinski AM (2004) The genome and proteome of coliphage T1. Virology 318:245–266
Fraser JS, Maxwell KL, Davidson AR (2007) Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr Opin Microbiol 10:382–387
Lehnherr H, Hansen AM, Ilyina T (1998) Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol Microbiol 30:454–457
Rodríguez-Rubio L, Martínez B, Donovan DM, Rodríguez A, García P (2012) Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. Crit Rev Microbiol [Epub ahead of print]
Piuri M, Hatfull GF (2006) A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol 62:1569–1585
Boulanger P, Jacquot P, Plançon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L (2008) Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem 283:13556–13564
Moak M, Molineux IJ (2004) Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol 51:1169–1183
Delbrück M (1940) The growth of bacteriophage and lysis of the host. J Gen Physiol 23:643–660
Abedon ST (2011) Lysis from without. Bacteriophage 1:46–49
Rodríguez L, Martínez B, Zhou Y, Rodríguez A, Donovan DM, García P (2011) Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol 11:138. doi:10.1186/1471-2180-11-138
Rashel M, Uchiyama J, Takemura I, Hoshiba H, Ujihara T, Takatsuji H, Honke K, Matsuzaki S (2008) Tail-associated structural protein gp61 of Staphylococcus aureus phage ΦMR11 has bifunctional lytic activity. FEMS Microbiol Lett 284:9–16
Rydman PS, Bamford DH (2000) Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol Microbiol 37:356–363
Bielen A, Cetković H, Long PF, Schwab H, Abramić M, Vujaklija D (2009) The SGNH-hydrolase of Streptomyces coelicolor has (aryl)esterase and a true lipase activity. Biochimie 91:390–400
Gutiérrez D, Martínez B, Rodríguez A, García P (2012) Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential. BMC Genomics 13:228. doi:10.1186/1471-2164-13-228
Cornelissen A, Ceyssens PJ, T’Syen J, Van Praet H, Noben JP, Shaburova OV, Krylov VN, Volckaert G, Lavigne R (2011) The T7-related Pseudomonas putida phage φ15 displays virion-associated biofilm degradation properties. PLoS One 6:e18597
Ma XX, Ito T, Chongtrakool P, Hiramatsu K (1985) Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J Clin Microbiol 44:4515–4527
Yamaguchi T, Hayashi T, Takami H, Nakasone K, Ohnishi M, Nakayama K, Yamada S, Komatsuzawa H, Sugai M (2000) Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol Microbiol 38:694–705
Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP (2010) The complete genomes of Staphylococcus aureus bacteriophages 80 and 80α–implications for the specificity of SaPI mobilization. Virology 407:381–390
Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, Wakiguchi H, Sugihara S, Sugiura T, Koda S, Muraoka A, Imai S (2003) Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis 187:613–624
Bae T, Baba T, Hiramatsu K, Schneewind O (2006) Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol 62:1035–1047
Hoshiba H, Uchiyama J, Kato S, Ujihara T, Muraoka A, Daibata M, Wakiguchi H, Matsuzaki S (2010) Isolation and characterization of a novel Staphylococcus aureus bacteriophage, phiMR25, and its therapeutic potential. Arch Virol 155:545–552
Daniel A, Bonnen PE, Fischetti VA (2007) First complete genome sequence of two Staphylococcus epidermidis bacteriophages. J Bacteriol 189:2086–2100
Swanson MM, Reavy B, Makarova KS, Cock PJ, Hopkins DW, Torrance L, Koonin EV, Taliansky M (2012) Novel bacteriophages containing a genome of another bacteriophage within their genomes. PLoS One 7:e40683
Acknowledgments
This research study was supported by grants AGL2012-40194-C02-01 (Ministry of Science and Innovation, Spain) and bacteriophage network FAGOMA. D. G. is a fellow of the Ministry of Science and Innovation, Spain. R. L., P. G. and A. R. are members of the FWO Vlaanderen–funded “Phagebiotics” research community (WO.022.09).
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material. Alignments of the staphylococcal phage genomes belonging to the family Siphoviridae, using the progressive-MAUVE software. Inside each block, nucleotide sequence similarity is indicated by the height of the coloured bars, while regions that are dissimilar are in white. (1) Comparison of genomes of members of “3alikevirus”; (2) MAUVE analysis of members of “77likevirus”; (3) “Phietalikevirus” subgroup 1; (4) “Phietalikevirus” subgroup 2; (5) “Phietalikevirus” subgroup 3.





Rights and permissions
About this article
Cite this article
Gutiérrez, D., Adriaenssens, E.M., Martínez, B. et al. Three proposed new bacteriophage genera of staphylococcal phages: “3alikevirus”, “77likevirus” and “Phietalikevirus”. Arch Virol 159, 389–398 (2014). https://doi.org/10.1007/s00705-013-1833-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1833-1