Abstract
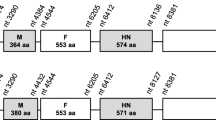
The complete DNA sequence of Marek’s disease virus (MDV) serotype 1 vaccine strain 814 was determined. It consisted of 172,541 bp, with an overall gene organization identical to that of the MDV-1 type strains. Comparative genomic analysis of vaccine strains (814 and CVI988) and other strains (CU-2, Md5, and Md11) showed that 814 was most similar to CVI988. Several unique insertions, deletions, and substitutions were identified in strain 814. Of note, a 177-bp insertion in the overlapping genes encoding the Meq, RLORF6, and 23-kDa proteins of strain 814 was identified, and a 69-bp deletion was also located in the origin of replication site (Ori) in the gene encoding RLORF12. Compared to the CVI988 vaccine strain, a deletion of 510 bp was identified in the UL36 gene. These analyses identified key mutations in the 814 strain and the vaccine strain that could be exploited for future MDV vaccine design.

Similar content being viewed by others
References
Roizman B (1982) In: Roizman B (ed) The Herpesviruses. Plenum, London, pp 1–23
Witter RL (1997) Increased virulence of Marek’s disease virus field isolates. Avian Dis 41:149–163
Lee LF, Wu P, Sui D, Ren D, Kamil J, Kung HJ, Witter RL (2000) The complete unique long sequence and the overall genomic organization of the GA strain of Marek’s disease virus. Proc Natl Acad Sci USA 97:6091–6096
Niikura M, Dodgson J, Cheng H (2005) Direct evidence of host genome acquisition by the alphaherpesvirus Marek’s disease virus. Arch Virol 151:537–549
Spatz SJ, Petherbridge L, Zhao Y, Nair V (2007) Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek’s disease virus. J Gen Virol 88:1080–1096
Spatz SJ, Zhao Y, Petherbridge L, Smith LP, Baigent SJ, Nair V (2007) Comparative sequence analysis of a highly oncogenic but horizontal spread-defective clone of Marek’s disease virus. Virus Genes 35:753–766
Tulman ER, Afonso CL, Lu Z, Zsak L, Rock DL, Kutish GF (2000) The genome of a very virulent Marek’s disease virus. J Virol 74:7980–7988
Spatz SJ, Rue CA (2008) Sequence determination of a mildly virulent strain (CU-2) of Gallid herpesvirus type 2 using 454 pyrosequencing. Virus Genes 36:479–489
Shamblin CE, Greene N, Arumugaswami V et al (2004) Comparative analysis of Marek’s disease virus (MDV) glycoprotein-, lytic antigen pp38- and transformation antigen Meq-encoding genes: association of Meq mutations with MDVs of high virulence. J Vet Microbiol 102:147–167
Jarosinski KW, Osterrieder N, Nair VK et al (2005) Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J Virol 79:11647–11659
Parcells MS, Lin SF, Dienglewicz RL et al (2001) Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J Virol 75(11):5159–5173
Fragnet L, Kut E, Rasschaert D (2005) Comparative functional study of the viral telomerase RNA based on natural mutations. J Biol Chem 280:23502–23515
Kamil JP, Tischer BK, Trapp S et al (2005) vLIP, a viral lipase homologue, is a virulrnce factor of Marek’s disease virus. J Virol 79(11):6984–6996
Spatz SJ, Silva RF (2007) Sequence determination of variable regions within the genomes of gallid herpesvirus-2 pathotypes. Arch Virol 152:1665–1678
Goldenberger D, Perschil I, Ritzler M, Altwegg M (1995) A simple “universal” DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. PCR Methods Appl 4:368–370
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings Bioinformatics 5:150–163
Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC (2002) Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071
Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ (2005) Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571
Zhao Y, Smith L, Petherbridge L, Baigent S, Nair V (2006) Investigation on the deubiquitinating enzyme activity of Marek’s Disease Virus UL36 homolog. In: Parcells MS, Morgan RW, Burnside J, Schmidt CJ, Bernberg EL, Eppler E (eds) 4th International Workshop on the Molecular Pathogenesis of Marek’s Disease Virus, p 27
Jones D, Lee L, Liu JL, Kung HJ, Tillotson JK (1992) Marek’s disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA 89:4042–4046
Chang KS, Ohashi K, Onuma M (2002) Suppression of transcription activity of the MEQ protein of oncogenic Marek’s disease virus serotype 1 (MDV1) by L-MEQ of non-oncogenic MDV1. J Vet Med Sci 64:1091–1095
Niikura M, Liu HC, Dodgson JB, Cheng HH (2004) A comprehensive screen for chicken proteins that interact with proteins unique to virulent strains of Marek’s disease virus. Poult Sci 83:1117–1123
Acknowledgments
This investigation was supported by the Special Fund of State Key Laboratory of China (No. SKLVBP200819) and Central Public-Interest Scientific Institution Basal Research Fund (No. ZGKJ200906), China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2011_1131_MOESM1_ESM.doc
Supplementary material 1 (DOC 31 kb). Supplementary Table S1 GenBank accession numbers for the genomic sequences of the seven MDV-1 strains used in the study
Rights and permissions
About this article
Cite this article
Zhang, F., Liu, CJ., Zhang, YP. et al. Comparative full-length sequence analysis of Marek’s disease virus vaccine strain 814. Arch Virol 157, 177–183 (2012). https://doi.org/10.1007/s00705-011-1131-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1131-8