Abstract
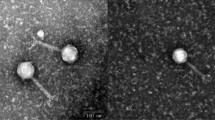
Bacteriophages contribute greatly to bacterial evolution. There has been limited investigation of enterococcal bacteriophages, and only two enterococcal bacteriophages have been sequenced completely. In this study, a novel enterococcal bacteriophage, EFRM31, was isolated from a piggery effluent sample and then characterized. The complete bacteriophage genome was determined by shotgun sequencing. EFRM31 belongs to the family Siphoviridae (order Caudovirales) and has a circular double-stranded DNA genome. The putative EFRM31 genome consists of 16945 nucleotides with a low GC content (34.5%) and does not contain CpG islands. The EFRM31 genome contains 82 putative open reading frames, including 17 with identities to genes required for the assembly of a head–tail bacteriophage and 6 hypothetical proteins of unknown function. In general, the sequencing results from EFRM31 revealed considerable similarity to another enterococcal bacteriophage, EFAP-1. This identity and the order of shared genes suggest a close relationship or a common ancestor for these two bacteriophages.

Similar content being viewed by others
References
Carey-Smith G, Billington C, Cornelius A, Hudson A, Heinemann J (2006) Isolation and characterisation of bacteriophages infecting Salmonella spp. FEMS Microbiol Lett 258:182–186
Chibani Azaiez SR, Fliss I, Simard RE, Moineau S (1998) Monoclonal antibodies raised against native major capsid proteins of lactococcal c2-like bacteriophages. Appl Environ Microbiol 64:4255–4259
Kaiser O, Bartels D, Bekel T, Goesmann A, Kespohl S, Puhler A, Meyer F (2003) Whole genome shotgun sequencing guided by bioinformatics pipelines—an optimized approach for an established technique. J Biotechnol 106:121–133
Lasken RS, Egholm M (2003) Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol 21:531–535
Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, Letellier G, Gilles M, Dethan G, Orlova EV, Couprie J, Tavares P, Zinn-Justin S (2009) Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA 106:8507–8512
Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW (2010) Novel bacteriophages in Enterococcus spp. Curr Microbiol 60:400–406
Ortmann AC, Lawrence JE, Sutlle CA (2002) Lysogeny and lytic viral production during a bloom of the Cyanobacterium synechococcus spp. Microb Ecol 43:225–231
Simjee S, Gill MJ (1997) Gene transfer, gentamicin resistance and enterococci. J Hosp Infect 36:249–259
Son JS, Jun SY, Kim EB, Park JE, Paik HR, Yoon SJ, Kang SH, Choi YJ (2009) Complete genome sequence of a newly isolated lytic bacteriophage, EFAP-1 of Enterococcus faecalis, and antibacterial activity of its endolysin EFAL-1. J Appl Microbiol 108:1769–1779
Uchiyama J, Rashel M, Maeda Y, Takemura I, Sugihara S, Akechi K, Muraoka A, Wakiguchi H, Matsuzaki S (2008) Isolation and characterization of a novel Enterococcus faecalis bacteriophage ΦEF24C as a therapeutic candidate. FEMS Microbiol Lett 278:200–206
Uchiyama J, Rashel M, Takemura I, Wakiguchi H, Matsuzaki S (2008) In silico and in vivo evaluation of bacteriophage ΦEF24C, a candidate for treatment of Enterococcus faecalis infections. Appl Environ Microbiol 74:4149–4163
Acknowledgments
We wish to thank Lynda Kutek, Sue Briggs, Karen Teague, Lora Mathews and Andrew Beck, School of Pharmacy and Medical Sciences, University of South Australia and Lyn Waterhouse, Adelaide Microscopy, University of Adelaide.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
GenBank accession number: GU815339.
Rights and permissions
About this article
Cite this article
Fard, R.M.N., Barton, M.D., Arthur, J.L. et al. Whole-genome sequencing and gene mapping of a newly isolated lytic enterococcal bacteriophage EFRM31. Arch Virol 155, 1887–1891 (2010). https://doi.org/10.1007/s00705-010-0800-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0800-3