Abstract
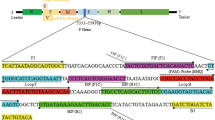
Five of 21 different Newcastle disease virus (NDV) strains isolated from China in 2007 gave false-negative results when detected with a previously reported loop-mediated isothermal amplification (LAMP) method, and mismatches were found between the target gene and LAMP primers. Therefore, an improved, sensitive, specific, and accelerated one-step reverse transcription (RT)-LAMP assay with degenerate primers was developed. Our data demonstrated that the improved RT-LAMP assay detected all 21 NDV isolates, had no cross-reaction with three other avian viruses, could be performed in 50 min less time, was 5-fold more sensitive than the previous LAMP assay, and achieved 96.8% sensitivity with 62 samples, including 30 field clinical samples, 24 experimentally infected samples, and 8 experimentally negative samples. Therefore, the improved RT-LAMP assay is a simple, rapid, and cost-effective method that is practical for less well-equipped laboratories and in the field.




Similar content being viewed by others
References
Alexander DJ (1997) Newcastle disease and other avian paramyxoviridae infections. In: Calnek BW, Barnes HJ, Beard CW et al (eds) Diseases of poultry, 10th edn. Iowa State University Press, Ames, pp 541–570
Alexander DJ (2004) Manual of diagnostic tests and vaccines for terrestrial animals, 5th edn. Office of International Des Epizooties, Paris, pp 270–282
Barbezange C, Jestin V (2002) Development of a RT-nested PCR test detecting pigeon Paramyxovirus-1 directly from organs of infected animals. J Virol Methods 106:197–207
Gohm DS, Thur B, Hofmann MA (2000) Detection of Newcastle disease virus in organs and faeces of experimentally infected chickens using RT-PCR. Avian Pathol 29:143–152
Kho CL, Mohd-Azmi ML, Arshad SS, Yusoff K (2000) Performance of an RT-nested PCR ELISA for detection of Newcastle disease virus. J Virol Methods 86:71–83
Liu XF, Wan HQ, Ni XX et al (2003) Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose in some regions of China during 1985–2001. Arch Virol 148:1387–1403
Lomniczi B, Wehmann E, Herczeg J et al (1998) Newcastle disease outbreaks in recent years in western Europe were caused by an old (virus isolation) and a novel genotype (VII). Arch Virol 143:49–64
Mayo MA (2002) A summary of taxonomic changes recently approved by ICTV. Arch Virol 147:1655–1663
Mori Y, Nagamine K, Tomita N, Notomi T (2001) Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun 289:150–154
Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 16:223–229
Nanthakumar T, Kataria RS, Tiwari AK et al (2000) Pathotyping of Newcastle disease virus by RT-PCR and restriction enzyme analysis. Vet Res Commun 24:275–286
Notomi T, Okayama H, Masubuchi H et al (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):E63
Pedersen JC, Senne DA, Woolcock PR et al (2004) Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002–2003 outbreak in California and other recent outbreaks in North America. J Clin Microbiol 42:2329–2334
Pham HM, Nakajima C, Ohashi K, Onuma M (2005) Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J Clin Microbiol 43:1646–1650
Seal BS, King DJ, Locke DP et al (1998) Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J Clin Microbiol 36:1141–1145
Wise MG, Suarez DL, Seal BS et al (2004) Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J Clin Microbiol 42:329–338
Acknowledgments
This work was supported by the grants from State Public Industry Scientific Research Programs (nyhyzx07-038) and Major Programs of Science Technology Strategic Plan (2007A020400006) of Guangdong, People's Republic of China. We thank Dr. George D. Liu for kindly revising the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, Q., Xue, C., Qin, J. et al. An improved reverse transcription loop-mediated isothermal amplification assay for sensitive and specific detection of Newcastle disease virus. Arch Virol 154, 1433–1440 (2009). https://doi.org/10.1007/s00705-009-0464-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0464-z