Abstract
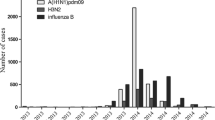
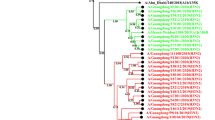
A total of 1,041 human influenza A virus isolates were collected at a clinic in Niigata, Japan, during eight influenza seasons from 2000 to 2007. The H3N2 subtype accounted for 75.4% of the isolates, and the rest were H1N1. Extremely high rates of amantadine-resistant strains of H3N2 subtype were observed in 2005/2006 (100%) and 2006/2007 (79.4%), while amantadine-resistant strains of H1N1 subtype were only detected in 2006/2007 (48.2%). Sequence and phylogenetic analysis of the HA1 subunit of the hemagglutinin (HA) gene revealed a characteristic linear trunk in the case of H3N2 viruses and a multi-furcated tree in the case of H1N1 and showed a higher sequence diversity among H3N2 strains than H1N1 strains. Mutations in the HA1 from both subtypes were mainly found in the globular region, and only one-third of these were retained for two or more successive years. Higher diversity of H3N2 viruses was mainly attributable to a higher fixation rate of non-synonymous mutations and to a lesser extent to a higher nucleotide substitution rate than for H1N1. Our analysis showed evidence of four positively selected sites in the HA1 of H1 and five sites in that of H3, four of which were novel. Finally, acquisition or loss of N-glycosylation sites was shown to contribute to the evolution of influenza A virus, especially in the case of H3N2, which had a higher tendency to acquire new glycosylation sites.


Similar content being viewed by others
References
Abe Y, Takashita E, Sugawara K, Matsuzaki Y, Muraki Y, Hongo S (2004) Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol 78:9605–9611
Belshe RB, Smith MH, Hall CB, Betts R, Hay AJ (1988) Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J Virol 62:1508–1512
Besselaar TG, Botha L, McAnerney JM, Schoub BD (2004) Antigenic and molecular analysis of influenza A (H3N2) virus strains isolated from a localised influenza outbreak in South Africa in 2003. J Med Virol 73:71–78
Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM (1999) Predicting the evolution of human influenza A. Science 286:1921–1925
Bush RM, Fitch WM, Bender CA, Cox NJ (1999) Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol 16:1457–1465
Carrat F, Flahault A (2007) Influenza vaccine: the challenge of antigenic drift. Vaccine 25:6852–6862
Caton AJ, Brownlee GG, Yewdell JW, Gerhard W (1982) The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427
CfD Control (1982) Concepts and procedures for laboratory based influenza surveillance. Center for Disease Control
Delano WL (2002) (www.pymol.org)
Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI (2007) Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis 196:249–257
Finkelman BS, Viboud C, Koelle K, Ferrari MJ, Bharti N, Grenfell BT (2007) Global patterns in seasonal activity of influenza A/H3N2, A/H1N1, and B from 1997 to 2005: viral coexistence and latitudinal gradients. PLoS ONE 2:e1296
Fitch WM, Leiter JM, Li XQ, Palese P (1991) Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci USA 88:4270–4274
Fitch WM, Bush RM, Bender CA, Cox NJ (1997) Long term trends in the evolution of H(3) HA1 human influenza type A. Proc Natl Acad Sci USA 94:7712–7718
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hay AJ, Zambon MC, Wolstenholme AJ, Skehel JJ, Smith MH (1986) Molecular basis of resistance of influenza A viruses to amantadine. J Antimicrob Chemother 18(Suppl B):19–29
Hayashida H, Toh H, Kikuno R, Miyata T (1985) Evolution of influenza virus genes. Mol Biol Evol 2:289–303
Hayden FG (2006) Antiviral resistance in influenza viruses—implications for management and pandemic response. N Engl J Med 354:785–788
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289
Jenkins GM, Rambaut A, Pybus OG, Holmes EC (2002) Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol 54:156–165
Knossow M, Skehel JJ (2006) Variation and infectivity neutralization in influenza. Immunology 119:1–7
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Li D, Saito R, Le MT, Nguyen HL, Suzuki Y, Shobugawa Y, Dinh DT, Hoang PV, Tran HT, Nghiem HK, Hoang LT, Huynh LP, Nguyen HT, Nishikawa M, Suzuki H (2008) Genetic analysis of influenza A/H3N2 and A/H1N1 viruses circulating in Vietnam from 2001 to 2006. J Clin Microbiol 46:399–405
Masuda H, Suzuki H, Oshitani H, Saito R, Kawasaki S, Nishikawa M, Satoh H (2000) Incidence of amantadine-resistant influenza A viruses in sentinel surveillance sites and nursing homes in Niigata, Japan. Microbiol Immunol 44:833–839
Nelson MI, Holmes EC (2007) The evolution of epidemic influenza. Nat Rev Genet 8:196–205
Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, George KS, Griesemer SB, Ghedin E, Sengamalay NA, Spiro DJ, Volkov I, Grenfell BT, Lipman DJ, Taubenberger JK, Holmes EC (2006) Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog 2:e125
Rambaut A (2000) Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395–399
Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC (2008) The genomic and epidemiological dynamics of human influenza A virus. Nature
Reece PA (2007) Neuraminidase inhibitor resistance in influenza viruses. J Med Virol 79:1577–1586
Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, Gregory V, Gust ID, Hampson AW, Hay AJ, Hurt AC, de Jong JC, Kelso A, Klimov AI, Kageyama T, Komadina N, Lapedes AS, Lin YP, Mosterin A, Obuchi M, Odagiri T, Osterhaus AD, Rimmelzwaan GF, Shaw MW, Skepner E, Stohr K, Tashiro M, Fouchier RA, Smith DJ (2008) The global circulation of seasonal influenza A (H3N2) viruses. Science 320:340–346
Saito R, Li D, Shimomura C, Masaki H, Le MQ, Nguyen HL, Nguyen HT, Phan TV, Nguyen TT, Sato M, Suzuki Y, Suzuki H (2006) An off-seasonal amantadine-resistant H3N2 influenza outbreak in Japan. Tohoku J Exp Med 210:21–27
Saito R, Li D, Suzuki Y, Sato I, Masaki H, Nishimura H, Kawashima T, Shirahige Y, Shimomura C, Asoh N, Degawa S, Ishikawa H, Sato M, Shobugawa Y, Suzuki H (2007) High prevalence of amantadine-resistance influenza a (H3N2) in six prefectures, Japan, in the 2005–2006 season. J Med Virol 79:1569–1576
Saito R, Suzuki Y, Li D, Zaraket H, Sato I, Masaki H, Kawashima T, Hibi S, Sano Y, Shobugawa Y, Oguma T, Suzuki H (2008) Increased incidence of adamantane-resistant influenza A(H1N1) and A(H3N2) viruses during the 2006–2007 influenza season in Japan. J Infect Dis 197:630–632; author reply 632–633
Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC (2007) The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 24:1811–1820
Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376
Stohr K (2002) Influenza–WHO cares. Lancet Infect Dis 2:517
Vigerust DJ, Ulett KB, Boyd KL, Madsen J, Hawgood S, McCullers JA (2007) N-linked glycosylation attenuates H3N2 influenza viruses. J Virol 81:8593–8600
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179
WHO (2005) Evolution of H5N1 avian influenza viruses in Asia. Emerg Infect Dis 11:1515–1521
Wiley DC, Wilson IA, Skehel JJ (1981) Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378
Wilson IA, Cox NJ (1990) Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 8:737–771
Wilson IA, Skehel JJ, Wiley DC (1981) Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366–373
Winter G, Fields S, Brownlee GG (1981) Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature 292:72–75
Wolf YI, Viboud C, Holmes EC, Koonin EV, Lipman DJ (2006) Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus. Biol Direct 1:34
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Wong WS, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ (2007) Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317:825–828
Zhang W, Jiang Q, Chen Y (2006) Evolution and variation of the H3 gene of influenza A virus and interation among hosts. Intervirology 50:287–295
Acknowledgments
This study was supported by Japan Grants-in-Aid for Scientific Research, from Monbu Kagakusho (Ministry of Education, Culture, Sports, Science and Technology, Japan), and Acute Respiratory Infections Panels, United States-Japan Cooperative Medical Science Program (US Department of Health and Human Services, US Department of State, United States, Ministry of Foreign Affairs, Ministry of Health, Labor, and Welfare, and Ministry of Education, Culture, Sports, Science and Technology, Japan). We are grateful to Dr. Akinori Miyashita and Dr. Ryozo Kuwano in the Department of Molecular Genetics, Bioresource Science Branch, Center for Bioresources, Brain Research Institute, Niigata University, for use of the DNA sequencer. We thank Ms. Akemi Watanabe for technical assistance for virus isolation, and Ms. Yoshiko Kato for intensive secretarial work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zaraket, H., Saito, R., Sato, I. et al. Molecular evolution of human influenza A viruses in a local area during eight influenza epidemics from 2000 to 2007. Arch Virol 154, 285–295 (2009). https://doi.org/10.1007/s00705-009-0309-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0309-9