Abstract
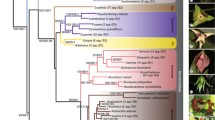
Studies of phylogenetic relationships among cypresses of the Old World (Cupressus; Cupressaceae) have been plagued by unresolved relationships, poor branch support, and conflict between data sets and methods of analysis. In this study, we combined 5.4 kb of aligned DNA sequence and 157 binary characters with previously published data in examining phylogenetic relationships among Cupressus species. Bayesian and parsimony analysis of the combined data or of the nuclear data alone always recovered three principal clades of Cupressus; however, tests of phylogenetic incongruence could not distinguish between competing relationships among the three principal Cupressus lineages. In contrast, incongruence tests often found statistically significant conflict between the nuclear and plastid data, particularly with respect to the placement of C. chengiana. Consistent with previous studies and prevailing taxonomic opinion, we find C. darjeelingensis more closely related to cypresses of the New World (Hesperocyparis). In contrast, we placed accessions of C. assamica and C. tonkinensis, two putatively Old World species suggested to be misidentified New World taxa by some authors, within well-supported Old World clades. Statistical analysis of genetic distances suggests instances in which taxa recognized as distinct species by some authors are identical or nearly so and may best be considered a single taxon. Conversely, we identify instances in which infraspecific taxa are more distantly related to one another than those traditionally recognized as distinct species. Factors confounding cypress taxonomies, including poor morphological differentiation, misidentification, and the use of accessions of questionable provenance, are discussed.




Similar content being viewed by others
References
Adams RP (2007) Juniperus maritima, the seaside juniper, a new species from Puget Sound, North America. Phytologia 89:263–283
Adams RP, Zanoni TA, Lara A, Barrero AF, Cool LG (1997) Comparisons among Cupressus arizonica Greene, C. benthamii Endl., C. lindleyi Klotz. ex Endl. and C. lusitanica Mill. using leaf essential oils and DNA fingerprinting. Essential Oil Res 9:303–309
Adams RP, Bartel JA, Price RA (2009) A new genus, Hesperocyparis, for the cypresses of the western Hemisphere (Cupressaceae). Phytologia 91:160–185
Averyanov LV, Hiep NT, Harder DK, Loc PK (2002) The history of discovery and natural habitats of Xanthocyparis vietnamensis (Cupressaceae). Turczananinowia 5:31–39
Bartel JA (2012) Hesperocyparis (western cypress). In: Baldwin BG, Goldman DH, Keil DJ, Patterson R, Rosatti TJ, Wilken DH (eds) The Jepson manual, vascular plants of California, 2nd edn. University of California Press, Berkeley, pp 136–138
Bartel JA, Adams RP, James SA, Mumba LE, Pandey RN (2003) Variation among Cupressus species from the western hemisphere based on random amplified polymorphic DNAs. Biochem Syst Ecol 31:693–702. https://doi.org/10.1016/s0305-1978(02)00229-6
Darlu P, Lecointre G (2002) When does the incongruence length difference test fail? Molec Biol Evol 19:432–437
Debreczy ZK, Musial K, Price R, Racz I (2009) Relationships and nomenclatural status of the nootka cypress (Callitropsis nootkatensis, Cupressaceae). Phytologia 91:140–159
Dolphin K, Belshaw R, Orme CD, Quicke DL (2000) Noise and incongruence: interpreting results of the incongruence length difference test. Molec Phylogen Evol 17(3):401–406. https://doi.org/10.1006/mpev.2000.0845
Dowton M, Austin AD (2002) Increased congruence does not necessarily indicate increased phylogenetic accuracy—the behavior of the incongruence length difference test in mixed model analyses. Syst Biol 51:19–31
Dyson WG, Herbin GA (1968) Studies on plant cuticular waxes-IV: leaf wax alkanes as a taxonomic discriminant for cypresses grown in Kenya. Phytochemistry 7:1339–1344
Farjon A (1993) Nomenclature of the Mexican cypress or “cedar of Goa”, Cupressus lusitanica Mill. (Cupressaceae). Taxon 42:81–84
Farjon A (1998) World checklist and bibliography of conifers. Royal Botanic Gardens, Kew
Farjon A (2005) A monograph of Cupressaceae and Sciadopitys. Royal Botanic Gardens, Kew
Farjon A (2010) A handbook of the world’s conifers, vol. 1. Brill Leiden, Boston
Farjon A, Hiep NT, Harder DK, Loc PK, Averyanov LV (2002) A new genus and species in the Cupressaceae (Coniferales) from northern Vietnam, Xanthocyparis vietnamensis. Novon 12:179–189
Farris JS, Kallersjo M, Kluge AG, Bult C (1995a) Constructing a significance test for incongruence. Syst Biol 44:570–572
Farris JS, Kallersjo M, Kluge AG, Bult C (1995b) Testing significance of incongruence. Cladistics 10:315–319
Franco JA (1945) A Cupressus lusitanica Miller, notas acera da sua história e sistemática. Agros (Lisbon) 28:3–27
Gadek P, Alpers DL, Heslewood MH, Quinn CJ (2000) Relationships within Cupressaceae sensu lato: a combined morphological and molecular approach. Amer J Bot 87:1044–1057
Goldman N, Anderson JP, Rodrigo AG (2000) Likelihood-based tests of topologies in phylogenetics. Syst Biol 49:652–670
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. https://doi.org/10.1080/10635150390235520
Herbin GA, Robins PA (1968) Studies on plant cuticular waxes-III: the leaf wax alkanes and hydroxy acids of some members of the Cupressaceae and Pinaceae. Phytochemistry 7:1325–1337
Hipp AL, Hall JC, Sytsma KJ (2004) Congruence versus phylogenetic accuracy: revisiting the incongruence length difference test. Syst Biol 53:81–89. https://doi.org/10.1080/10635150490264752
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066
Kim ST, Donoghue MJ (2008) Incongruence between cpDNA and nrITS trees indicates extensive hybridization within Eupersicaria (Polygonaceae). Amer J Bot 95(9):1122–1135. https://doi.org/10.3732/ajb.0700008
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Molec Evol 29:170–179. https://doi.org/10.1007/bf02100115
Little EL (1966) Varietal transfers in Cupressus and Chamaecyparis. Madroño 18:161–167
Little EL (1970) Names of New World cypresses (Cupressus). Phytologia 20:429–445
Little DP (2005) Evolution and circumscription of the true cypresses (Cupressaceae: Cupressus and Callitropsis): A combined molecular and morphological approach. Cornell University, Ithaca
Little DP (2006) Evolution and circumscription of the true Cypresses (Cupressaceae: Cupressus). Syst Bot 31:461–480
Little DP, Schwarzbach AE, Adams RP, Hsieh C-F (2004) The circumscription and phylogenetic relationships of Callitropsis and the newly described genus Xanthocyparis (Cupressaceae). Amer J Bot 91:1871–1880
Maerki D (2013) Which Latin name for the Tsenden? Bull Cupressus Conserv Proj 2:39–71
Maerki D (2014) Appendix: taxonomic index of the genus Cupressus. Bull Cupressus Conserv Proj 3:48
Maerki D (2017) Note on Cupressus assamica Silba. Bull Cupressus Conserv Proj 6:74–76
Mao K, Hao G, Liu J, Adams RP, Milne RI (2010) Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. New Phytol 188:254–272. https://doi.org/10.1111/j.1469-8137.2010.03351.x
Martinez M (1947) Los Cupressus de Mexico. Ann Inst Biol 18:71–149
Muller K (2005) SeqState: primer design and sequence statistics for phylogenetic DNA data sets. Appl Bioinform 4:65–69
Muller K (2006) Incorporating information from length-mutational events into phylogenetic analysis. Molec Phylogen Evol 38:667–676. https://doi.org/10.1016/j.ympev.2005.07.011
Neng Z (1980) Species nova generis Cupressi. Acta Phytotaxon Sin 18:210
Planet PJ (2006) Tree disagreement: measuring and testing incongruence in phylogenies. J Biomed Inform 39:86–102. https://doi.org/10.1016/j.jbi.2005.08.008
Posada D (2008) jModelTest: phylogenetic model averaging. Molec Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Qu X-J, Jin J-J, Chaw S-M, Li D-Z, Yi T-S (2017) Multiple measures could alleviate long branch attraction in phylogenomic reconstruction of Cupressoideae (Cupressaceae). Sci Rep 7:41005. https://doi.org/10.1038/srep41005
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Rushforth K, Adams RP, Zhong M, X-q Ma, Pandey RN (2003) Variation among Cupressus species from the eastern hemisphere based on Random Amplified Polymorphic DNAs (RAPDs). Biochem Syst Ecol 31:17–24
Sambrook J, Russell DW (2001) Molecular cloning, a laboratory manual, vol. 3. Cold Spring Harbor Laboratory Press, Cold Springs Harbor, New York
Shaw J, Lickey EB, Beck JT, Sb Farmer, Liu W, Miller J, Siripun KC, Winde CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer J Bot 92:142–166
Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508. https://doi.org/10.1080/10635150290069913
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molec Biol Evol 16:1114–1116
Silba J (1981) Revised generic concepts of Cupressus L. (Cupressaceae). Phytologia 49:390–399
Silba J (1982) Addendum to a revision of Cupressus L. (Cupressaceae). Phytologia 52:349–361
Silba J (1984) An international census of the Coniferae, I. Phytologia Mem. H. N. & A. L. Moldenke, Plainfield
Silba J (1986) Encyclopeaedia Coniferae. Phytologia Mem.,VIII. H. N. & A. L. Moldenke, Plainfield
Silba J (1988) A new species of Cupressus L. from Tibet (Cupressaceae). Phytologia 65:333336
Silba J (1990) A supplement to the international census of the Coniferae, II. Phytologia 68:7–78
Silba J (1994) The trans-pacific relationship in Cupressus in India and North America. J Int Conifer Preserv Soc 1:1–28
Silba J (1998) A monograph of the genus Cupressus L. J Int Conifer Preserv Soc 5:1–98
Silba J (2005) A monograph of the genus Cupressus L. in the twenty-first century. J Int Conifer Preserv Soc 12:30–101
Silba J (2006) The chronological history and taxonomic variation of the genus Cupressus (Cupressaceae) in India. Acta Bot Yunnanica 28:469–470
Silba J (2013) Authentic documentation on seedling embryology of Cupressus torulosa from natural populations in India. Bull Cupressus Conserv Proj 2:26–31
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Templeton AR (1983) Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37:221–224
Terry RG, Adams RP (2015) A molecular re-examination of phylogenetic relationships among Juniperus, Cupressus, and the Hesperocyparis–Callitropsis–Xanthocyparis clades of Cupressaceae. Phytologia 97:66–74
Terry RG, Bartel JA, Adams RP (2012) Phylogenetic relationships among the New World cypresses (Hesperocyparis; Cupressaceae): evidence from noncoding chloroplast DNA sequences. Pl Syst Evol 298:1987–2000. https://doi.org/10.1007/s00606-012-0696-3
Terry RG, Pyne MI, Bartel JA, Adams RP (2016) A molecular biogeography of the New World cypresses (Callitropsis, Hesperocyparis; Cupressaceae). Pl Syst Evol 302:921–942. https://doi.org/10.1007/s00606-016-1308-4
Wan-Chun C, Li-Kuo F, Ching-Yung C (1975) Gymnospermae sinicae. Acta Phytotaxon Sin 13:56–123
Wolf CB (1948) The New World cypresses. Aliso 1:1–250
Xiang Q, Li J (2005) Derivation of Xanthocyparis and Juniperus from within Cupressus: evidence from sequences of the nrDNA internal transcribed spacer region. Harvard Pap Bot 9:375–382
Xu T, Abbott RJ, Milne RI, Mao K, Du FK, Wu G, Ciren Z, Miehe G, Liu J (2010) Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evol Biol 10:194. https://doi.org/10.1186/1471-2148-10-194
Yang ZY, Ran JH, Wang XQ (2012) Three genome-based phylogeny of Cupressaceae s.l.: further evidence for the evolution of gymnosperms and Southern Hemisphere biogeography. Molec Phylogen Evol 64:452–470. https://doi.org/10.1016/j.ympev.2012.05.004
Yani A, Baradat P, Bernard-Dagan C (1990) Chemotaxinomy [sic] of Cupressus species. In: Progress in EEC Research on Cypress Diseases, Agrimed Research Programme. Commission of the European Communities, Agriculture, Report EUR 12493 EN:29-38
Yoder AD, Irwin JA, Payseur BA (2001) Failure of the ILD to determine data combinability for slow loris phylogeny. Syst Biol 50:408–424
Zhao N (1980) Species nova generis Cupressi. Acta Phytotaxon Sin 18:210
Acknowledgements
We thank Bob Adams for providing leaf tissue and helpful comments on the manuscript. We also thank an anonymous reviewer for many comments helpful in improving the manuscript. This work was funded in part by a Research Enhancement Grant awarded to RGT through the College of Arts and Sciences, Lamar University. Support from the Lamar University Department of Biology is gratefully acknowledged.
Funding
This study was funded by a Research Enhancement (REG2011) from Lamar University to RGT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Jürg Schönenberger.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Combined nuclear and chloroplast DNA data with binary characters in Nexus format.
Online Resource 2. Nuclear or chloroplast DNA data (depending on exclusion set) with binary characters in Nexus format.
Online Resource 3. Nuclear and chloroplast DNA data with binary characters used in distance analyses.
Online Resource 4. Voucher and source information for outgroups and a listing of outgroup and non-ingroup taxa used in the various analyses presented here.
Online Resource 5. Features of the parsimony analyses and trees presented in this study.
Online Resource 6. Output from analysis of the nuclear data.
Online Resource 7. Output from analysis of the nuclear and chloroplast data combined.
Online Resource 8. Topologies used to generate constraints, the analyses constrained, and results from parsimony-based and maximum likelihood-based tests of topological incongruence.
Online Resource 9. Output from analysis of the chloroplast data.
Rights and permissions
About this article
Cite this article
Terry, R.G., Schwarzbach, A.E. & Bartel, J.A. A molecular phylogeny of the Old World cypresses (Cupressus: Cupressaceae): evidence from nuclear and chloroplast DNA sequences. Plant Syst Evol 304, 1181–1197 (2018). https://doi.org/10.1007/s00606-018-1540-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-018-1540-1