Abstract
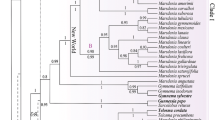
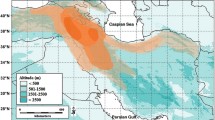
Allium section Rhizirideum constitutes a group of approximately 24 species, distributed mostly in steppe areas of the Eurasian temperate zone. Its phylogenetic relationships are therefore relevant for understanding of the evolutionary and biogeographical patterns of the Eurasian steppes. Based upon DNA sequences from two plastid regions (trnQ-rps16 and trnL-rpl32) and the internal transcribed spacer region of nuclear ribosomal DNA, the phylogenetic relationships in Allium section Rhizirideum are investigated by using maximum parsimony and Bayesian inference. Dated phylogenies revealed (1) that diversification started in the upper Pliocene and further speciation events occurred during the Pleistocene and (2) a clear division of the section Rhizirideum into an “Asiatic” and “European” geographical group. Nomenclature, distribution maps and identification key for all species are provided. Origin and diversification within this section thus reflect the development and history of the modern Eurasian steppe.





Similar content being viewed by others
References
Agapova ND, Arkharova KB, Vakhtina LI, Zemskova EA, Tarvis LV (1990) Numeri chromosomatum magnoliophytorum florae URSS: Aceraceae—Menyanthaceae. Nauka, Sectio Leninopolitana, St Petersburg
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Bardy KE, Albach DC, Schneeweiss GM, Fischer MA, Schönswetter P (2010) Disentangling phylogeography, polyploid evolution and taxonomy of a woodland herb (Veronica chamaedrys group, Plantaginaceae s.l.) in southeastern Europe. Molec Phylogen Evol 57:771–786
Barkalov VY (1987) Sem. 143. Lukovye – Alliaceae J. Agardh. In: Charkevicz SS (ed) Plantae vasculares Orientis Extremi Sovietici, Tomus 2. Nauka, Leningrad, pp 376–393
Blattner FR (1999) Direct amplification of the entire ITS region from poorly preserved plant material using recombinant PCR. Biotechniques 27:1180–1186
Bolkhovskikh ZV, Grif VG, Zakhar’eva OI, Matveyeva TS (1969) Khromosomnye chisla tsvetkovych rastenii. Nauka, Leningrad
Cheremushkina VA (2004) Biologiia lukov Evrazii, Novosibirsk
Choi HJ (2015) Portrayal of Allium spurium G. Don (Amaryllidaceae) from the border area of China and North Korea: a putative unrecorded species in the Korean Peninsula. Botanica. Pacifica 4:1–3. doi:10.17581/bp.2015.04202
Choi HJ, Oh BU (2010) A new species and a new combination of Allium sect. Rhizirideum (Alliaceae) from northeastern China and Korea. Brittonia 62:199–205
Delplancke M, Alvarez N, Benoit L, Espindola A, Joly HI, Neuenschwander S, Arrigo N (2013) Evolutionary history of almond tree domestication in the Mediterranean basin. Molec Ecol 22:1092–1104
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Ehrendorfer F (1980) Chapter 3 polyploidy and distribution. In: Lewis WH (ed) Polyploidy, biological relevance. Plenum Press, New York, pp 45–60
Favarger C (1961) Sur l’emploides nombres de chromosomes en géographie botanique historique. Ber Geobot Inst ETH Stiftung Rübel 32:119–146
Favarger C (1984) Cytogeography and biosystematics. In: Grant WF (ed) Plant biosystematics. Academic Press, Vancouver, pp 453–476
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Franzke A, Hurka H, Janssen D, Neuffer B, Friesen N, Markov M (2004) Molecular signals for late Tertiary/Early Quarternary range splits of an Eurasian steppe plant: Clausia aprica (Brassicaceae). Molec Ecol 13:2789–2795
Friesen N (1987) Rod Allium L. In: Malyshev LI, Peshkova GA (eds) Flora Sibiri. Araceae-Orchidaceae, Novosibirsk, pp 55–96
Friesen N (1988) Lukovye Sibiri: sistematika, kariologiia, khorologiia. Nauka-Sibirskoe otd, Novosibirsk
Friesen N (1992) Systematics of the Siberian polyploid complex in subgenus Rhizirideum (Allium). In: Hanelt P, Hammer K, Knupffer H (eds) The genus Allium: taxonomic problems and genetic resources, proceedings of an international symposium held at Gatersleben. Institut für Pflanzengenetik und Kulturpflanzenforschung, Gatersleben, pp 55–66
Friesen N (1995) The genus Allium L. in the flora of Mongolia. Feddes Repert 106:59–81
Friesen N, Hermann N (1998) Taxonomy, chorology and evolution of Allium lusitanicum: the European “A. senescens”. Linzer Biol Beitr 30:815–830
Friesen N, Bolognini G, Nimis PL (1993) Quantitative phytogeography of the genus Allium in Siberia and Mongolia. Nordic J Bot 5:295–307
Friesen N, Fritsch RM, Blattner FR (2006) Phylogeny and intrageneric classification of Allium L. (Alliaceae) based on nuclear ribosomal DNA ITS. Aliso 22:372–395
Friesen N, German DA, Hurka H, Herden T, Oyuntsetseg B, Neuffer B (2015) Dated phylogenies and historical biogeography of Dontostemon and Clausia (Brassicaceae) mirror the palaegeographical history of the Eurasian steppe. J Biogeogr. doi:10.1111/jbi.12658
Fritsch RM (2001) Taxonomy of the genus Allium: contributions from IPK Gatersleben. Herbertia 56:19–50
Fritsch RM, Friesen N (2002) Evolution, domestication, and taxonomy. In: Rabinovich HD, Currah L (eds) Allium crop science: recent advances. CABI Publishing, Wallingford, pp 5–30
Herden T, Hanelt P, Friesen N (2016) Phylogeny of Allium L. subgenus Anguinum (G. Don. ex W. D. J. Koch) N. Friesen (Amaryllidaceae). Molec Phylogen Evol 95:79–93
Hermy M, Honnay O, Firbank L, Grasof-Bokdam C, Lawesson JE (1999) An ecological comparison between ancient and other forest plant species of Europe, and the implications for forest conservation. Biol Conservation 91:9–22
Hewitt GM (2001) Speciation, hybrid zones and phylogeography: or seeing genes in space and time. Molec Ecol 10(3):537–549
Huang C-C, Hung K-H, Wang W-K, Ho C-W, Huang C-L, Hsu T-W, Osada N et al (2012) Evolutionary rates of commonly used nuclear and organelle markers of Arabidopsis relatives (Brassicaceae). Gene 499:194–201
Hurka H, Friesen N, German DA, Franzke A, Neuffer B (2012) ‘Missing link’ species Capsella orientalis and Capsella thracica elucidate evolution of model plant genus Capsella (Brassicaceae). Molec Ecol 21:1223–1238
Huson DH, Scornavacca C (2010) A survey of combinatorial methods for phylogenetic networks. Genome Biol Evol 3:23–35. doi:10.1093/gbe/evq077
Huson DH, Scornavacca C (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61:1061–1067. doi:10.1093/sysbio/sys062
IPCN (1979–2016) Index to plant chromosome numbers. Goldblatt P, Johnson DE (eds) Missouri Botanical Garden, St. Louis. http://mobot.mobot.org/W3T/Search/ipcn.html. Accessed 15 Jan 2016
Jeanmogin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Kamelin RV (2004) Lektsii po sistematike rastenii. Glavy teoreticheskoi sistematiki rastenii, Izdatel’stvo “AzBuka”, Barnaul
Kay K, Whittall J, Hodges S (2006) A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol Biol 6:36
Kirschner J, Kirschnerova L, Stĕpánek J (2007) Generally accepted plant names based on material from the Czech Republic and published in 1753–1820. Preslia 79:323–365
Koldaeva MN (2015) On the infraspecific diversity of Allium spirale Willd. ex Schlecht. (Alliaceae) in the Russian Far East. Turczaninowia 18:5–10
Krogulevich RE, Rostovtseva TS (1984) Khromosomnye chisla tsvetkovykh rastenii Sibiri i Dal’nego Vostoka. Nauka, Sibirskoe Otdelenie, Novosibirsk
Li QQ, Zhou DZ, He XH, Xu Y, Zhang YC, Wei XQ (2010) Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann Bot (Oxford) 106:709–733
Nürk NM, Uribe-Convers S, Gehrke B, Tank DC, Blattner FR (2015) Oligocene niche shift, Miocene diversification: cold tolerance and accelerated speciation rates in the St. John’s Worts (Hypericum, Hypericaceae). BMC Evol Biol 15:80. doi:10.1186/s12862-015-0359-4
Parisod C, Holderegger R, Brochmann C (2010) Evolutionary consequences of autopolyploidy. New Phytol 186:5–17
Pastor J (1982) Karyology of Allium species from the Iberian Peninsula. Phyton (Horn) 22:171–200
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Ronquist R, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Schmitt T (2007) Molecular biogeography of Europe: pleistocene cycles and postglacial trends. Front Zool 4:11
Seregin AP, Anaćkov G, Friesen N (2015) Molecular and morphological revision of the Allium saxatile group (Amaryllidaceae): geographical isolation as the driving force of underestimated speciation. Bot J Linn Soc 178:67–101. doi:10.1111/boj.12269
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequence to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Amer J Bot 94:275–288
Sinitsyna TA, Friesen N (2008) Phylogeny of the Allium L. section Rhizirideum G. Don ex W. D. J. Koch based on molecular-genetic data: In: Proceedings of 7th international scientific-practical conference on problems of botany of South Siberia and Mongolia. (Barnaul, 21–24 Oktober 2008): 323–326
Stehlik I, Blattner FR, Holderegger R, Bachmann K (2002) Nunatak survival of the high Alpine plant Eritrichium nanum (L.) Gaudin in the central Alps during the ice ages. Molec Ecol 11:2027–2036
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (* and other methods), vers. 4.0. Sinauer Associates, Inc., Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl Molec Biol 17:1105–1109
Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F (1998) Comparative phylogeography and postglacial colonization routes in Europe. Molec Ecol 7:453–464
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molec Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Vvedensky AI (1935) Rod 267. Luk: Allium L. In: Komarov VL (ed) Flora URSS IV, Izdatel’stvo AN SSSR, Leningrad, pp 112–280
Xu JM, Kamelin RV (2000) Allium L. In: Wu ZY, Raven PH (eds) Flora of China. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis
Acknowledgments
Curators and managers of the visited herbaria are greatly acknowledged for help and providing leaf material. We thank Herbert Hurka and Reinhard Fritsch for helpful comments on the manuscript and Lucille Schmieding for correcting the English. We want to give special thanks to the reviewers, for their valuable comments and suggestions on this manuscript. For financial support, we are grateful to DAAD for a Grant (PKZ A/07/90071) to the first author as well as to DFG for supporting the collecting of Allium specimen in Siberia and Mongolia for N. Friesen and to RSF (Russian Science Foundation, Project 14-14-00472) for T. Sinitsyna.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We confirm that this work is original and has not been published elsewhere nor is it currently under consideration for publication elsewhere. We the authors declare that the manuscript “Dater phylogeny and biogeography of the eurasian section rhizirideum G.Don ex W.D.J.Koch (Allium L., Amaryllidaceae)” (PLSY-D-16-00066) complies with all the requirements of “the ethical responsibilities of the authors” part in the “instruction for authors” guideline (Cope guidlines).
Additional information
Handling editor: Livia Wanntorp.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Online resources ESM1
Sources of geographical information. (PDF 371 kb)
Online resources ESM2
Chromosome numbers of Allium sect. Rhizirideum species and sources of information. (PDF 298 kb)
Online resources ESM3
ITS Alignment of all accessions of section Rhizirideum. (PDF 451 kb)
Online resources ESM4
ITS Alignment with only diploid species of section Rhizirideum. (PDF 418 kb)
Online resources ESM5
Plastid Alignment. (PDF 376 kb)
Online resources ESM6
Hybridogenic Network, based on ITS and plastid trees. (PDF 721 kb)
Online resources ESM7
Estimated mean ages. (PDF 375 kb)
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online resource 1. Sources of geographical information.
Online resource 2. Chromosome numbers of Allium sect. Rhizirideum species and sources of information.
Online resource 3. ITS Alignment of all accessions of section Rhizirideum.
Online resource 4. ITS Alignment with only diploid species of section Rhizirideum.
Online resource 5. Plastid Alignment.
Online resource 6. Hybridogenic Network, based on ITS and plastid trees.
Online resource 7. Estimated mean ages.
Rights and permissions
About this article
Cite this article
Sinitsyna, T.A., Herden, T. & Friesen, N. Dated phylogeny and biogeography of the Eurasian Allium section Rhizirideum (Amaryllidaceae). Plant Syst Evol 302, 1311–1328 (2016). https://doi.org/10.1007/s00606-016-1333-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-016-1333-3