Abstract
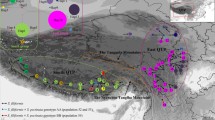
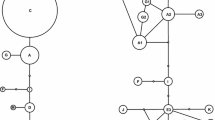
The Hengduan Mountains is the core of the Sino-Himalayan Floristic Region. Different views have been postulated from phylogeographic studies in understanding the historical biological events shaping present-day distributions and species diversities in this area. We analyzed the phylogeography of Rosa soulieana, a widely distributed rose species in dry and semi-dry valleys of the Hengduan Mountains of southwestern China using two cpDNA intergenic spacers. 9 cpDNA haplotypes were confirmed in 650 individuals from 39 populations, with the most dominant one existing in 36 populations. The relationship among the cpDNA haplotypes did not reflect the morphology-based intra-specific taxonomy of different varieties. There was very low cpDNA genetic diversity and relatively high population differentiation combined with lack of phylogeographic structure. The populations from Northwestern Yunnan, Southeastern Tibet and upper Yalongjiang River embraced most of the species genetic diversity, but with contribution of different historical effects. Northwestern Yunnan and upper Yalongjiang River appear to be the species refugia and centers of adaptive diversification, while Southeastern Tibet rather represents the species ‘melting pot’ in the Quaternary climate oscillations.




Similar content being viewed by others
References
Avise JC (2004) Molecular markers, Natural History, and Evolution, 2nd edn. Sinauer Associates, Sunderland
Birky CW Jr (1988) Evolution and variation in plant chloroplast and mitochondrial genomes. In: Gottlieb LD, Jain SK (eds) Plant evolutionary biology. Chapman & Hall, London, pp 23–53
Chen SY, Wu GL, Zhang DJ, Gao QB, Duan YZ, Zhang FQ, Chen SL (2008) Potential refugium on the Qinghai-Tibet Plateau revealed by the chloroplast DNA phylogeography of the alpine species Metagentiana striata (Gentianaceae). Bot J Linn Soc 157:125–140
Cheng J, Liu XQ, Gao ZJ, Tang DX, Yue JW (2001) Effect of the Tibetan plateau uplifting on geological environment of the Yunnan Plateau. Geoscience 15:290–296
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Cun YZ, Wang XQ (2010) Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Molec Phylogenet Evol 56:972–982
De Cock K, Vander Mijnsbrugge K, Breyne P, Van Bockstaele E, Van Slycken J (2008) Morphological and AFLP-based Differentiation within the Taxonomical Complex Section Caninae (subgenus Rosa). Ann Bot (Oxford) 102:685–697
Dumolin-Lapegue S, Pemonge MH, Petit RJ (1997) An enlarged set of consensus primers for the study of organelle DNA in plants. Molec Ecol 6:393–397
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Molec Ecol 11:2571–2581
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plants conservation. Annual Rev Ecol Syst 24:217–242
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gao LM, Möller M, Zhang XM, Hollingsworth ML, Liu J, Mill RR, Gibby M, Li DZ (2007) High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam. Molec Ecol 16:4684–4698
Ge XJ, Zhang LB, Yuan YM, Hao G, Chiang TY (2005) Strong genetic differentiation of the East-Himalayan Megacodon stylophorus (Gentianaceae) detected by inter-simple sequence repeats (ISSR). Biodivers Conserv 14:849–861
Graur D, Li WH (2000) Fundamentals of molecular evolution, 2nd edn. Sinauer Associates, Sunderland
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hamilton MB (1999) Four primer pairs for the amplification of chloroplastic intragenic regions with intraspecific variation. Molec Ecol 8:513–525
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos T Roy Soc B 351:1291–1298
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequence of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci 359:183–195
Kato S, Iwata H, Tsumura Y, Mukai Y (2011) Genetic structure of island populations of Prunus lannesiana var. speciosa revealed by chloroplast DNA, AFLP and nuclear SSR loci analyses. J Pl Res 124:11–23
Kim ST, Donoghue M (2008) Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low-copy nuclear region. Proc Natl Acad Sci USA 105:12370–12375
Kozak KH, Wiens JJ (2006) Does niche conservatism promote speciation? A case study in North American salamanders. Evolution 60:2604–2621
Ku TC (1985) Rosa. Flora Reipublicae Popularis Sinicae, vol 37. Science Press, Beijing, pp 360–455
Ku TC, Robertson KR (2003) Rosa (Rosaceae). In: Wu ZY, Raven PH (eds) Flora of China, vol 9. Science Press, Beijing; Missouri Botanical Garden Press, St. Louis, pp 339–381
Li JJ, Wen SX, Zhang QS, Wang FB, Zheng BX, Li BY (1979) A discussion on the period, amplitude and type of the uplift of the Qinghai-Xizang Plateu. Sci Sin 22:1314–1328
Li Y, Stocks M, Hemmilä S, Källman T, Zhu HT, Zhou YF, Chen J, Liu JQ, Lascoux M (2010) Demographic histories of four spruce (Picea) species of the Qinghai-Tibetan Plateau and neighboring areas inferred from multiple nuclear loci. Molec Biol Evol 27:1001–1014
Li Y, Zhai SN, Qiu YX, Guo YP, Ge XJ, Comes HP (2011) Glacial survival east and west of the ‘Mekong–Salween-Divide’ in the Himalaya-Hengduan Mountains region as revealed by AFLP and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Molec Phylogenet Evol 59:412–424
Li GD, Yue LL, Sun H, Qian ZG (2012) Phylogeography of Cyananthus delavayi (Campanulaceae) in Hengduan Mountains inferred from variation in nuclear and chloroplast DNA sequences. J Syst Evol 50:305–315
Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu XY (2012) Phylogeographic studies of plants in China: advances in the past and directions in the future. J Syst Evol 50:267–275
Liu J, Sun P, Zheng XY, Potter D, Li KM, Hu CY, Teng YW (2013) Genetic structure and phylogeography of Pyrus pashia (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genom 9:433–441
Liu JQ, Duan YW, Hao G, Ge XJ, Sun H (2014) Evolutionary history and underlying adaptation of alpine plants on the Qinghai-Tibet Plateau. J Syst Evol 52(3):241–249
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annual Rev Ecol Syst 15:65–95
Molina-Montenegro MA, Atala C, Gianoli E (2010) Phenotypic plasticity and performance of Taraxacum officinale (dandelion) in habitats of contrasting environmental heterogeneity. Biol Invasions 12:2277–2284
Müller-Schärer H, Fischer M (2001) Genetic structure of the annual weed Senecio vulgaris in relation to habitat type and population size. Molec Ecol 10:17–28
Nei M (1987) Molecular Evolutionary Genetics. Columbia University Press, New York
Nei M, Tajima F (1983) Maximum likelihood estimation of the number of nucleotide substitutions for restriction sites data. Genetics 105:207–216
Newton AC, Allnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999) Molecular phytogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
Opgenoorth L, Vendramin GG, Mao K, Miehe G, Miehe S, Liepelt S, Liu J, Ziegenhagen B (2010) Tree endurance on the Tibetan Plateau marks the world’s highest known tree line of the Last Glacial Maximum. New Phytol 185:332–342
Panchal M (2007) The automation of nested clade phylogeographic analysis. Bioinformatics 23:509–510
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Aparajita M, Müller-Starck G, Demesure-Musch B, Palmé A, Martín JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molec Ecol 14:689–701
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Buckley T (2004) Model selection and model averaging in phylogenetics: advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Molec Phylogenet Evol 59:225–244
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molec Biol Evol 9:552–569
Royden LH, Burchfiel BC, van der Hilst RD (2008) The geological evolution of the Tibetan Plateau. Science 321:1054–1058
Rozas J, Sanchez-Del Barrie JC, Messeguer X, Rozas R (2003) DNASP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Rusanov K, Kovacheva N, Vosman B, Zhang L, Rajapakse S, Atanassov A, Atanassov I (2005) Microsatellite analysis of Rosa damascena Mill. accessions reveals genetic similarity between genotypes used for rose oil production and old Damask rose varieties. Theor Appl Genet 111:804–809
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Molec Ecol 7:465–474
Soltis PS, Gitzendanner MA (1999) Molecular systematics and the conservation of rare species. Conserv Biol 13:471–483
Sun HL, Zheng D (1998) Formation/evolution and development of Qinghai-Xizang (Tibetan) Plateau. Guangdong Science and Technology Press, Guangzhou
Sun YS, Wang AL, Wan DS, Wang Q, Liu JQ (2012) Rapid radiation of Rheum (Polygonaceae) and parallel evolution of morphological traits. Molec Phylogenet Evol 63:150–158
Swofford DL (2000) PAUP4.0b10. Phylogenetic Analysis Using Parsimony and Other Methods, vol 4. Sinauer Associates, Sunderland
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molec Biol Evol 28:2731–2739
Templetion AR, Sing CF (1993) A cladistics analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics 134:659–669
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vosman B, Visser D, Rouppe J, van der Voort, Smulders MJM, van Eeuwijk F (2004) Detection of mutants and the establishment of essential derivation among rose varieties using AFLP. Theor Appl Genet 109:1718–1725
Wang FY, Gong X, Hu CM, Hao G (2008) Phylogeography of an alpine species Primula secundiflora inferred from the chloroplast DNA sequence variation. J Syst Evol 46:13–22
Wang LY, Abbott RJ, Zheng W, Chen P, Wang YJ, Liu JQ (2009) History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Molec Ecol 18:709–721
Wang JF, Pan YZ, Gong X, Chiang YC, Kuroda C (2011) Chloroplast DNA variation and phylogeography of Ligularia tongolensis (Asteraceae), a species endemic to the Hengduan Mountains Region of China. J Syst Evol 49:108–119
Webb T, Bartlein PJ (1992) Global changes during the last 3 million years: climatic controls and biotic response. Annual Rev Ecol Syst 23:141–173
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wu ZY (1988) The Hengduan Mountain Flora and her significance. J Jap Bot 63:297–311
Wu XY, Chen M, Wang QG, Zhou NN, Zhang T, Yan HJ, Qiu XQ, LI SB, Zhang H, Jian HY, Tang KX (2014) Comparative study on the breeding systems of Rosa praelucens and Rosa soulieana. Acta Hortic Sin 41:2075–2084
Xia T, Chen SL, Chen SY, Ge XJ (2005) Genetic variation within and among populations of Rhodiola alsia (Crassulaceae) native to the Tibetan Plateau as detected by ISSR markers. Biochem Genet 43:87–101
Xu TT, Abbott RJ, Milne RI, Mao K, Du F-K, Wu GL, Ciren ZX, Miehe G, Liu JQ (2010) Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evol Biol 10:194
Yang FS, Li YF, Ding X, Wang XQ (2008) Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with Quaternary climate change. Molec Ecol 17:5135–5145
Yue XK, Yue JP, Yang LE, Li ZM, Sun H (2011) Systematics of the genus Salweenia (Leguminosae) from Southwest China with discovery of a second species. Taxon 60:1366–1374
Yue LL, Chen G, Sun WB, Sun H (2012) Phylogeography of Buddleja crispa (Buddlejaceae) and its correlation with drainage system evolution in southwestern China. Amer J Bot 99:1726–1735
Zhang TC, Sun H (2011) Phylogeographic structure of Terminalia franchetii (Combretaceae) in southwest China and its implications for drainage geological history. J Pl Res 124:63–73
Zhang YH, Volis S, Sun H (2010) Chloroplast phylogeny and phylogeography of Stellera chamaejasme on the Qinghai-Tibet Plateau and in adjacent regions. Molec Phylogenet Evol 57:1162–1172
Zhang TC, Comes HP, Sun H (2011) Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture event. Molec Phylogenet Evol 60:1–12
Zhang FQ, Gao QB, Zhang DJ, Duan YZ, Li YH, Fu PC, Xing R, Gulzar K, Chen SL (2012) Phylogeography of Spiraea alpine (Rosaceae) in the Qinghai-Tibetan Plateau inferred from chloroplast DNA sequence variations. J Syst Evol 50:276–283
Zhang L, Yan HF, Wu W, Yu H, Ge XJ (2013) Comparative transcriptome analysis and marker development of two closely related primrose species (Primula poissonii and Primula wilsonii). BMC Genom 14:329
Zhou SZ, Wang XL, Wang J, Xu LB (2006) A preliminary study on timing of the oldest Pleistocene glaciation in Qinghai-Tibetan Plateau. Quatern Int 154–155:44–51
Zhou ZQ, Bao WK, Wu FZ, Wu N (2009) Capability and limitation of regeneration of Rosa hugonis and Rosa soulieana in the dry valley of the upper Minjiang River. Acta Ecol Sin 29:1931–1939
Acknowledgments
We sincerely thank Dr. Sergei Volis for comments and suggestions to improve the manuscript. We thank Dr. Yong-Hong Zhang for helping in data analysis. We thank Dr. Hong-Guang Zha and Ms. Min-Shu Song for assistance with the molecular techniques. We are also grateful to Dr. Lu-lin Ma, Ms. Xian-Qin Qiu and Mr. Shu-bin Li for helping with some field survey. This study was supported by a Joint Project between National Natural Science Foundation of China and Yunnan Natural Science Foundation (Grant No. U1136601), the 100 Talents Program of the Chinese Academy of Sciences (No. 2011312D11022) to Prof. Hang Sun; the National Natural Science Foundation of China 31260198 to Ms. Hong-Ying Jian, and the Academic and Technical Talents Training Project of Yunnan Province (Grant No. 2013 HB092) to Ms. Hong-Ying Jian. We are also thankful to two anonymous reviewers for their valuable comments in improving the quality of the present manuscript.
Conflict of interest
The authors of this article have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Yunpeng Zhao.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jian, HY., Tang, KX. & Sun, H. Phylogeography of Rosa soulieana (Rosaceae) in the Hengduan Mountains: refugia and ‘melting’ pots in the Quaternary climate oscillations. Plant Syst Evol 301, 1819–1830 (2015). https://doi.org/10.1007/s00606-015-1195-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-015-1195-0