Abstract
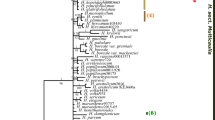
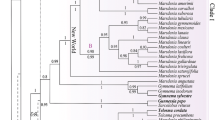
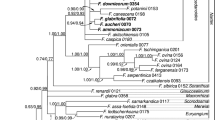
This study represents the molecular phylogeny of the tribe Hedysareae using nrDNA ITS and plastid trnL-F and matK sequence data. The molecular analyses united nine Hedysaroid genera in a well-supported monophyletic group (tribe Hedysareae). The tribe is circumscribed here as comprising Alhagi, Corethrodendron, Ebenus, Eversmannia, Hedysarum, Onobrychis, Sulla, Taverniera, and Greuteria. Our analyses revealed that Caragana and allies form a well-supported clade, the so-called Caraganean clade, which is weakly allied with the Hedysareae. Within the Hedysareae, Alhagi is the first diverging lineage followed by Sulla clade and a large assemblage of the other Hedysaroid genera. Ebenus and Taverniera were monophyletic. All analyses showed that Hedysarum is not monophyletic in the present circumscription. Most members of the genus with the inclusion of Sartoria hedysaroides were gathered in a single clade. The two other Hedysarum species (H. membranaceum and H. argyreum) were sister taxa and placed in a clade with Eversmannia subspinosa and Corethrodendron. Onobrychis was retrieved as a weakly supported clade. On the basis of the present sequence data together with non-molecular characteristics, we transferred H. aculeolatum to Sulla and S. hedysaroides to Hedysarum as well as elevated H. membranaceum and H. argyreum to the new generic rank named as Greuteria Amirahmadi & Kaz. Osaloo. A diagnostic key to the genera of the tribe is provided.





Similar content being viewed by others
References
Ahangarian S, Kazempour Osaloo S, Maassoumi AA (2007) Molecular phylogeny of the tribe Hedysareae with special reference to Onobrychis (Fabaceae) as inferred from nrDNA ITS sequences. Iran Jour Bot 13:64–74
Aksoy H, Unal F, Aytac Z (2001) Karyological study on four endemic Ebenus L. Taxa (Fabaceae) in Turkey. Caryologica 54:307–311
Ali SI (1977) Papilionaceae. In: Nasir E, Ali SI (eds) Flora of West Pakistan, no. 100. University of Karachi, Karachi
Amirahmadi A, Kazempour Osaloo S, Maassoumi AA (2010) Loss of chloroplast trnLUAA intron in two species of Hedysarum (Fabaceae): evolutionary implications. IJB 8:150–155
Archie JW (1989) A randomization test for phylogenetic information in systematic data. Syst Zool 38:239–252
Arslan E, Ertuğrul K (2010) Genetic relationships of the genera Onobrychis, Hedysarum, and Sartoria using seed storage proteins. Turk J Biol 34:1–7
Arslan E, Ertuğrul K, Tugay O, Dural H (2012) Karyological studies of the genus Onobrychis Mill. and the related genera Hedysarum L. and Sartoria Boiss. & Heldr. (Fabaceae, Hedysareae) from Turkey. Caryologia 65:11–17
Awmack CS, Lock JM (2002) The genus Alhagi (Leguminosae: Papilionoideae) in the Middle East. Kew Bull 57:435–443
Aytaç Z (2000) The genus Ebenus L. (Leguminosae/Fabaceae) in Turkey. Karaca Arbor Mag 5:145–171
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacer of nuclear ribosomal DNA in plants: an example from the Compositae. Mol Phylogenet Evol 1:3–16
Baldwin BJ, Sanderson MJ, Markprter J, Wojciechowski MF, Campbell C, Donoghue JM (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence of Angiosperm phylogeny. Ann Miss Bot Gard 82:247–277
Basiner TFJ (1845) Enumeratio monographica specierum generis Hedysari. Mém Acad Imp Sci St Pétersbourg 6:45–97
Battandier JA (1888) Dicotylédones–Thalamiflores. In: Battandier JA, Trabut LC (eds) Flore de ľ Algérie, Fascicule 1. Typographie Adolphe Jourdan, Alger
Bentham G (1853) On three new genera connected with the Indian flora. Hooker’s J Bot Kew Gard Misc 5:304–309
Bentham G (1865) Leguminosae. In: Bentham G, Hooker JD (eds) Genera plantarum, vol 1, part 2. L. Reeve & Co., London, pp 434–600
Boissier E (1856) Diagnoses plantarum Orietalium Novarum. Ser. II, vol 5. Herrmann, Lipsiae, p 92
Chamberlain DF (1970) Alhagi. In: Davis PH (ed) Flora of Turkey and the East Aegean Island, vol 3. Edinburgh University Press, Edinburg, pp 596–597
Chase MW, Cowan RS, Hollingsworth PM et al (2007) A proposal for a standardised protocol to barcode all land plants. Taxon 56:295–299
Chennaoui H, Marghali S, Marrakchi M, Trifi- Farah N (2007) Phylogenetic relationships in the North African genus Hedysarum as inferred from ITS sequences of nuclear ribosomal DNA. Genet Resour Crop Evol 54:389–397
Chennaoui-Kourda H, Marghali M, Marrakchi M, Trifi-Farah N (2007) genetic diversity of Sulla genus (Hedysareae) and related species using inter-simple sequence repeat (ISSR) markers. Biochem Syst Ecol 35:682–688
Chennaoui-Kourda H, Marghali S, Zitouna N, Trifi-Farah N (2012) Phylogenetic relationships of Mediterranean Hedysarea species assessed by AFLP markers. Plant Syst Evol 298:51–58
Choi BH, Ohashi H (1996) Pollen morphology and taxonomy Hedysarum and and its related genera of the tribe Hedysareae (Leguminosae—Papilinoideae). J Jap Bot 71:191–213
Choi BH, Ohashi H (1998) Proposal to conserve the name Hedysarum (Leguminosae: Papilionoideae) with a conserved type. Taxon 47:977
Choi BH, Ohashi H (2003) Generic criteria and an infrageneric system for Hedysarum and related genera (Papilinoideae–Leguminosae). Taxon 52:567–576
Choi BH, T Nemato, Ohashi H (1999) Anatomy of nodal regions and leaves in Hedysarum and related genera (Leguminosae). J Jap Bot 74:236–250
Chrtková-Žertová A (1968) Hedysarum L. In: Tutin TG et al (eds) Flora Europaea, vol 2. Cambridge University Press, Cambridge, pp 185–187
Cui H (1998) Flora Reipublicae Popularis Sinicae, vol 42. Science Press, Beijing
Cunningham CW (1997) Can three incongruence tests predict when data should be combined? Mol Biol Evol 14:733–740
De Candolle AP (1825) Prodromus systematis naturalis regni vegetabilis, vol 2. Treuttel and Wurtz, Paris, p 307
Douzery E, Pridgeon A, Kores P, Linder HP, Kurzweil H, Chase M (1999) Molecular phylogenetics of Diseae (Orchidaceae): a contribution from nuclear ribosomal ITS sequences. Amer Jour Bot 86:887–899
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Edgar RC (2004) Muscle: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797
Ems SC, Morden CW, Dixon CK, Wolfe KH, dePamphilis CW, Palmer JD (1995) Transcription, splicing and editing of plastid RNAs in the non-photosynthetic plant Epifagus virginiana. Plant Mol Biol 29:721–733
Farris JS, Kallersjo M, Kluge AG, Bult C (1995) Testing significance of incongruence. Cladistics 10:315–319
Fedtschenko BA (1902) Generis Hedysari revisio. Acta Horti Petrop 19:183–342
Fedtschenko BA (1972) Hedysarum L. (Leguminosae). In: Komarov VL, Shishkin BK, Bobrov EG (eds) Flora of the USSR, vol 13. Israel Program for Scientific Translation, Jerusalem, pp 199–243
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 38:783–791
Goldblatt P (1981) Cytology and the phylogeny of Leguminosae. In: Polhill RM, Raven PH (eds) Advances in legume systematics, part 2. Royal Botanic Gardens, Kew, pp 427–463
Gorshkova SG (1972) Eversmannia Bge. (Leguminosae). In: Komarov VL, Shishkin BK, Bobrov EG (eds) Flora of the USSR, vol 13. Israel Program for Scientific Translation, Jerusalem, pp 198–199
Greuter W, Raus T (1986) Med-Checklist Notulae 13. Willdenowia 16:103–116
Grossheim AA (1972) Onobrychis Adans. (Leguminosae). In: Komarov VL, Shishkin BK, Bobrov EG (eds) Flora of the USSR, vol 13. Israel Program for Scientific Translation, Jerusalem, pp 244–281
Hall TA (1999) BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hedge IC (1970) Hedysarum, Onobrychis and Sartoria. In: Davis PH (ed) Flora of Turkey and the East Aegean Islands, vol 3. Edinburgh University Press, Edinburgh, pp 549–590
Hilu KW, Liang H (1997) The matK gene: sequence variation and application in plant systematics. Am J Bot 84:830–839
Hilu KW, Borsch T, Muller K, Soltis DE, Soltis PS, Savolainen V, Chase MW et al (2003) Angiosperm phylogeny based on matK sequence information. Am J Bot 90:1758–1776
Huber-Morath A (1970) Ebenus L. In: Davis PH (ed) Flora of Turkey and the East Aegean Islands, vol 3. Edinburgh University Press, Edinburgh, pp 590–596
Hutchinson J (1964) The genera of flowering plants, vol 1. Oxford University Press, Oxford
Javanmardi F, Kazempour Osaloo S, Maassoumi AA, Nejadsattari T (2012) Molecular phylogeny of Astragalus section Alopecuroidei (Fabaceae) and its allies based on nrDNA ITS and three cpDNAs, matK, trnT-trnYand trnH-psbA sequences. Biochem Syst Ecol 45:171–178
Johnson LA, Soltis DE (1995) Phylogenetic inference using matK sequences. Ann Mo Bot Gard 82:149–175
Kazemi M, Kazempour Osaloo S, Maassoumi AA, Rastegar Pouyani E (2009) Molecular phylogeny of selected Old World Astragalus (Fabaceae): incongruence among chloroplast trnL, ndhF and nuclear ribosomal DNA ITS sequences. Nord J Bot 27:425–436
Kazempour Osaloo S, Kawano S (1999) Molecular systematics of Trilliaceae II. Phylogenetic analyses of Trillium and its allies using sequences of rbcL and matK genes of cpDNA and internal transcribed spacers of 18S-26S nr DNA. Plant Spec Biol 14:75–94
Kazempour Osaloo S, Utech FH, Ohara M, Kawano S (1999) Molecular systematics of Trilliaceae I. Phylogenetic analyses of Trillium using matK gene sequences. J Plant Res 112:35–49
Kazempour Osaloo S, Maassoumi AA, Murakami N (2005) Molecular systematics of the Old World Astragalus (Fabaceae) as inferred from nrDNA ITS sequence data. Brittonia 57:367–381
Kenicer G, Kajita T, Pennington R, Murata J (2005) Systematics and biogeography of Lathyrus (Leguminoseae) based on internal transcribed spacer and cpDNA sequence data. Am J Bot 92:1199–1209
Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. PNAS USA 105:2923–2928
Lewis G, Schrire B, Mackinder B, Lock M (2005) Legumes of the world. Royal Botanical Gardens, Kew
Linnaeus C (1763) Species plantarum, vol 2, 2nd edn. Impensis Laurentii Salvii, Holmiae
Liu Y, Xu LR, Chang Z, Zhu X, Sun H, Yakovlev GP, Choi BH, Larsen K, Bartholomew B (2010) Tribe Hedysareae (Fabaceae) In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, vol 10. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, pp 512–545
Lock JM (2005) Tribe Hedysarae. In: Lewis G, Schrire B, Mackinder B, Lock M (eds) Legumes of the world. Royal Botanical Gardens, Kew, pp 489–495
Lock JM, Simpson K (1991) Legume of the West Asia: a check-list. Royal Botanical Gardens, Kew
Mabberley DJ (2008) The plant-book. A portable dictionary of the higher plants, 3rd edn. Cambridge University Press, Cambridge
Maire R (1939) Contribution á ľétude de la Flore de ľAfrique du Nord. Bull Soc Hist Nat Afrique N 30:327–370
Meikle RD (1977) Flora of Cyprus, vol 1. Bentham–Moxon Trust, Royal Botanic Gardens
Mozaffarian V (1988) New species and new plant records from Iran. Iran J Bot 4:61–70
Neuhaus H, Link G (1987) The chloroplast tRNALYS(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a matures related polypeptide. Curr Genet 11:251–257
Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008) Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Res 8:480–490
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Oskoueiyan R, Kazempour Osaloo S, Maassoumi AA, Nejadsattari T, Mozaffarian V (2010) Phylogenetic status of Vavilovia formosa (Fabaceae–Fabeae) based on nrDNA ITS and cpDNA sequences. Biochem Syst Ecol 38:313–319
Page DM (2001) Treeview (Win32) version 1.6.6. Available at. http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
Plunkett GM, Downie SR (1999) Major lineages within Apiaceae subfamily Apioideae: a comparison of chloroplast restriction site and DNA sequence data. Am J Bot 86:1014–1026
Polhill RM (1981) Hedysareae. In: Polhil RM, Rave PH (eds) Advances in legume systematics, part 1. Royal Botanic Gardens, Kew, pp 367–370
Polhill RM, Raven PH (eds) (1981) Advances in legume systematics, part 1. Royal Botanic Gardens, Kew, pp 1–425
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Sys Biol 53:793–808
Qiu YL, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW (1999) The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 402:404–407
Ragupathy S, Newmaster SG, Murugesan M, Balasubramaniam V (2009) DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the hill tribes of the Western Ghats in southern India. Mol Ecol Resour 9:164–171
Ranjbar M, Karamian R (2003) Caraganeae, a newtribe with notes on the genus Chesneya Lindel. (Fabaceae) from flora of the Iran. Thaiszia J Bot 13:67–75
Rechinger KH (1984) Tribus Hedysareae (Papilionaceae II). In: Rechinger KH (ed) Flora Iranica, no. 157. Akademische Druck, Graz, pp 365–475
Riahi M, Zarre S, Maassoumi AA, Kazempour Osaloo S, Wojciechowski MF (2011) Towards a phylogeny for Astragalus section Caprini (Fabaceae) and its allies based on nuclear and plastid DNA sequences. Plant Syst Evol 293:119–133
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1–210
Safaei Chaei Kar S, Ghanavati F, Mozafari J, Naghavi MR, Amirabadizadeh H, Darvish F (2012) Phylogenetic relationships of Onobrychis Mill. (Fabaceae: Papilionoideae) based on ITS sequences of nuclear ribosomal DNA and morphological traits. Crop Breed J 2:91–99
Sanderson MJ, Wojciechowski MF (1996) Diversification rates in a temperate legume clade: “are there so many species” of Astragalus (Fabaceae). Am J Bot 83:1488–1502
Sang T, Crawford DJ, Stuessy T (1995) Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implication for biogeography and concerted evolution. Proc Natl Acad Sci USA 92:6813–6817
Selvaraj D, Sarma RK, Sathishkuman R (2008) Phylogenetic analysis of chloroplast matK gene from Zingiberaceae for plant DNA barcoding. Bioinformation 3:24–27
Shaperenko KK (1972) Alhagi Adans. (Leguminosae). In: Komarov VL, Shishkin BK, Bobrov EG (eds) Flora of the USSR, vol 13. Israel Program for Scientific Translation, Jerusalem, pp 281–285
Shaw J, Lickey E, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The Tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:42–166
Silvestro D, Michalak I (2011) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337
Širjaev G (1925) Onobrychis generis revisio critica. Pars prima. Publications Faculte des Sciences de l’ Université Masaryk 56:1–197
Soltis DE, Kuzoff RK, Conti E, Gornall R, Ferguson K (1996) matK and rbcL gene sequence data indicate that Saxifraga (Saxifragaceae) is polyphyletic. Am J Bot 83:371–382
Starr JR, Naczi RFC, Chouinard BN (2009) Plant DNA barcodes and species resolution in sedges (Carex, Cyperaceae). Mol Ecol Resour 9:151–163
Steele KP, Vilgalys R (1994) Phylogenetic analyses of Polemoniaceae using nucleotide sequences of the Plastid gene matK. Syst Bot 19:126–142
Steele KP, Wojciechowski MF (2003) Phylogenetic analyses of tribes Trifolieae and Vicieae based on sequences of the plastid gene matK (Papilionoideae: Leguminosae). In: Klitgaard BB, Bruneau A (eds) Advances in legume systematics, part 10. Higher level systematics. Royal Botanic Gardens, Kew, pp 355–370
Stirton CH (2005) Tribe Psoraleeae. In: Lewis G, Schrire B, Mackinder B, Lock M (eds) Legumes of the world. Royal Botanical Gardens, Kew, pp 447–451
Sugita M, Shinozaki K, Sugiura M (1985) Tobacco chloroplast tRNALys(UUU) gene contains a 2.5-kilobase-pair intron: an open reading frame and a conserved boundary sequence in the intron. P N A S USA 82:3557–3561
Sun H, Bartholomew B (2010) Eversmannia (Fabaceae–Hedysareae). In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, vol 10. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, pp 526
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sinauer Associates, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal Primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Thulin M (1985) Revision of Taverniera (Leguminosae: Papilionoideae). Symb Bot Ups 25:44–94
Townsend CC (1974) Leguminales. In: Townsend CC, Guest ER (eds) Flora of Iraq, vol 3. Ministry of Agriculture and Agrarian Reform of the Republic of Iraq, Republic of Iraq
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis DH (ed) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wojciechowski MF (2003) Reconstructing the phylogeny of legumes (Leguminosae): an early 21st century perspective. In: Klitgaard B, Bruneau A (eds) Advances in legume systematics, part 10. Royal Botanic Gardens, Kew, pp 5–35
Wojciechowski MF (2005) Astragalus (Fabaceae): a molecular phylogenetic perspective. Brittonia 57:382–399
Wojciechowski MF, Sanderson MJ, Hu JM (1999) Evidence on the monophyly of Astragalus (Fabaceae) and its major subgroups based on nuclear ribosomal DNA ITS and chloroplast DNA trnL intron data. Syst Bot 24:409–437
Wojciechowski MF, Sanderson MJ, Steele KP, Liston A (2000) Molecular phylogeny of the “temperate herbaceous tribes” of papilionoid legumes: a super tree approach. In: Herendeen P, Bruneau A (eds) Advances in legume systematics, part 9. Royal Botanic Garden, Kew, pp 277–298
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of Legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91:1846–1862
Xu LR, Choi BH (2010a) Corethrodendron (Fabaceae–Hedysareae) In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, vol 10. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, pp 512–514
Xu LR, Choi BH (2010b) Hedysarum (Fabaceae–Hedysareae). In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, vol 10. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, pp 514–525
Yakolev GP, Sytin AK, Roskov YR (1996) Legumes of Northern Eurasia: a check-list. Royal Botanic Gardens, Kew
Yakovelov GP (1979) Notes on the taxonomy of the genus Alhagi Gagneb (Fabaceae). Bot Zhurn (Mosc Leningr) 64:1794–1799
Yildiz B, Ciplak B, Aktoklu E (1999) Fruit morphology of sections of the genus Onobrychis Miller (Fabaceae) and its phylogenetic implications. Isr J Plant Sci 47:269–282
Yu WB, Huang PH, Ree RH, Liu ML, Li DZ, Wang H (2011) DNA barcoding of Pedicularis L. (Orobanchaceae): evaluating four universal barcode loci in a large and hemiparasitic genus. J Syst Evol 49:425–437
Zwickl D (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas at Austin, USA
Acknowledgments
The present study was financially supported in part by Grant-in-Aids for Scientific Research, No. 89002433, to S. K. O (corresponding author) from INSF (Iran National Science Foundation). This work represents partial fulfillment of the requirement for obtaining a PhD degree by the first author from Tarbiat Modares University. We would like to thank the staff of the herbaria of ANK, FUMH, G, GAZI, MSB, TARI, TUH, KNRCH, and WANRCH to allow studying herbarium specimens and providing leaf materials. We would like to thank the anonymous reviewers for their comments on the improving the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Amirahmadi, A., Kazempour Osaloo, S., Moein, F. et al. Molecular systematics of the tribe Hedysareae (Fabaceae) based on nrDNA ITS and plastid trnL-F and matK sequences. Plant Syst Evol 300, 729–747 (2014). https://doi.org/10.1007/s00606-013-0916-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0916-5