Abstract
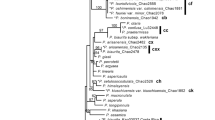
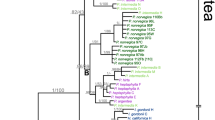
Hybridization is particularly likely to occur in initial and young polyploid complexes, and interspecies hybridization between diverged species usually leads to a complicated reticulate evolution. The Ranunculus cantoniensis complex and its allied species include R. chinensis (2x), R. silerifolius var. silerifolius (2x), R. cantoniensis (4x), R. trigonus (2x), R. shuichengensis (2x), R. diffusus (4x), R. repens (4x), R. vaginatus (5x) and R. sieboldii (6x, 8x). Many morphological intermediates can be found between the members of this complex, and the relationship among members is complicated. By analyzing internal transcribed spacers and nrDNA FISH (fluorescence in situ hybridization) signals, we unraveled the phylogenetic and genetic constitution of the various taxonomic units of this complex. Haplotypes were highly separated by median-joining network analysis and at least four haplogroups emerged in which there were 11 primary haplotypes; six out of ten taxa shared haplotype 1, suggesting that haplotype 1, a variation of the primary haplotype R. chinensis, served as the pivotal genome in the complex. The pollen characteristics and electrophoretic patterns of R. vaginatus (5x) showed it to be an intermediate between R. diffusus (4x) and R. sieboldii (6x). The distribution of R. vaginatus (5x) was located at the junction of the distributions of R. diffusus (4x) and R. sieboldii (6x). Ranunculus vaginatus (5x) shared haplotypes 7 and 8 with R. diffusus (4x), and haplotypes 8 and 9 with R. sieboldii (6x). This proved that R. vaginatus (5x) emerged from hybridization between R. diffusus (4x) and R. sieboldii (6x). The results of FISH also support a hybrid origin of R. vaginatus (5x). The findings of this study clearly show that there are only eight taxa in this polyploid complex including R. chinensis (2x), R. silerifolius var. silerifolius (2x), R. trigonus (2x), R. silerifolius var. dolicanthus(2x), R. cantoniensis (4x), R. diffusus (2x), R. vaginatus (5x) and R. sieboldii (6x, 8x). These taxa associated with each other by hybridizing with the pivotal genome. Ranunculus cantoniensis (4x) and R. vaginatus (5x) arose from hybridization events between diverged species in the polyploid complex, leading to a complicated reticulate evolution.





Similar content being viewed by others
Abbreviations
- CTAB:
-
Cetyltrimethyl ammonium bromide
- FISH:
-
Fluorescence in situ hybridization
- ITS:
-
Internal transcribed spacer
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16(1):37–48
De Wet J (1971) Polyploidy and evolution in plants. Taxon 20(1):29–35
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19(1):11–15
Fuertes AJ, Nieto FG (2003) Additive polymorphisms and reticulation in an ITS phylogeny of thrifts (Armeria, Plumbaginaceae). Mol Phylogenet Evol 28(3):430–447
Fujishima H, Kurita M (1975) Chromosome studies in Ranunculaceae, XXVI. Variation in karyotype of Ranunculus ternatus var. glaber. Mem Ehime Univ Sci Ser B 7(3):62–68
Gagnidze RJ, Churadze MV (1984) Chromosome numbers in some species of the section chrysanthe of the genus Ranunculus (Ranunculaceae) from Georgia. Bot Zhurn 69(11):1570–1571
Guggisberg A, Mansion G, Conti E (2009) Disentangling reticulate evolution in an Arctic-Alpine polyploid complex. Syst Biol 58(1):55–73
Gulyás G, Sramkó G, Molnár AV, Rudnóy S, Illyés Z, Balázs T, Bratek Z (2005) Nuclear ribosomal DNA ITS paralogs as evidence of recent interspecific hybridization in the genus Ophrys (Orchidaceae). Acta Biol Cracov Bot 47:61–67
Hizume M (1993) Chromosomal localization of 5S rRNA genes in Vicia faba and Crepis capillaris. Cytologia 58(4):417–421
John C, Templeton AR (1994) Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol Phylogenet Evol 3(2):102–113
Liao L, Xu LL (1996) A study on pollen morphology of Sect. Ranunculus from China. Acta Bot Boreal Occident Sin 16(3):255–261 (Chinese)
Liao L, Xu LL (1997) New taxa of the genus Ranunculus from China and their karyotypes. Acta Phytotaxon Sin 35(1):57–62 (Chinese)
Liao L, Xu LL, Chen Y, Fang L (1995) Studies of karyotypes of Ranunculus cantoniensis polyploid complex and its allied species. Guihaia 33(3):230–239 (Chinese)
Liao L, Xu LL, Zhang DM, Fang L, Deng HS, Shi JW, Li TJ (2008) Multiple hybridization origin of Ranunculus cantoniensis (4x): evidence from trnL-F and ITS sequences and fluorescent in situ hybridization (FISH). Plant Syst Evol 276(1):31–37
Linder CR, Rieseberg LH (2004) Reconstructing patterns of reticulate evolution in plants. Am J Bot 91(10):1700–1708
Lo EYY, Stefanović S, Dickinson TA (2010) Reconstructing reticulation history in a phylogenetic framework and the potential of allopatric speciation driven by polyploidy in an agamic complex in crataegus (Rosaceae). Evolution 64(12):3593–3608
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16(6):562
Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F (2009) TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25(1):126
Nitta JH, Ebihara A, Ito M (2011) Reticulate evolution in the Crepidomanes minutum species complex (Hymenophyllaceae). Am J Bot 98(11):1782–1800
Okada H (1977) Chromosome variations in Ranunculus quelpaertensis and its allied species. J Jpn Bot 52:360–369
Okada H (1981) On sexual isolation caused by karyotype variations in Ranunculus silerifolius Lév. J Jpn Bot 56:41–49
Okada H (1984) Polyphyletic allopolyploid origin of Ranunculus cantoniensis (4x) from R. silerifolius (2x) × R. chinensis (2x). Plant Syst Evol 148(1):89–102
Okada H (1989) Cytogenetical changes of offsprings from the induced tetraploid hybrid between Ranunculus silerifolius (2n = 16) and R. chinensis (2n = 16) (Ranunculaceae). Plant Syst Evol 167(3):129–136
Poczai P, Hyvönen J (2010) Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Mol Biol Rep 37(4):1897–1912
Rieseberg LH (2001) Chromosomal rearrangements and speciation. Trends Ecol Evol 16(7):351–358
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15(2):174–175
Sang T, Crawford DJ, Stuessy TF (1995) Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. PNAS 92(15):6813
Soltis DE, Soltis PS, Rieseberg DLH (1993) Molecular data and the dynamic nature of polyploidy. Crit Rev Plant Sci 12(3):243–273
Soltis DE, Soltis PS, Tate JA (2004) Advances in the study of polyploidy since plant speciation. New Phytol 161(1):173–191
Stebbins GL (1971) Secondary modifications of polyploids. In: Barrington EJW, Willis J (eds) Chromosomal evolution in higher plants. Edward Arnold, London, pp 147–154
Suh Y, Thien LB, Reeve HE, Zimmer EA (1993) Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of ribosomal DNA in Winteraceae. Am J Bot 80:1042–1055
Takahashi C (2003) Physical mapping of rDNA sequences in four karyotypes of Ranunculus silerifolius (Ranunculaceae). J Plant Res 116(4):331–336
Tamura M (1978) Ranunculus cantoniensis group in Japan. J Geobot 26:34–40
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882
Troitsky AV, Melekhovets YF, Rakhimova GM, Bobrova VK, Valiejo-Roman KM, Antonov AS (1991) Angiosperm origin and early stages of seed plant evolution deduced from rRNA sequence comparisons. J Mol Evol 32(3):253–261
Wang WT (1995a) A revision of the genus Ranunculus in China (I). Bull Bot Res 15(2):137–180 (Chinese)
Wang WT (1995b) A revision of the genus Ranunculus in China (II). Bull Bot Res 15:275–329 (Chinese)
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfond DH, Sninsky JJ, White TJ (eds) PCR protocols a guide to methods and applications. Academic Press, New York, pp 315–322
Xu LL, Liao L, Fang L, Sun GL (1997) A study on peroxidase isozymes of Ranunculus cantoniensis DC. Complex and its allied species. J Wuhan Bot Res 15(1):43–48 (Chinese)
Zhang DM, Sang T (1998) Chromosomal structural rearrangement of Paeonia brownii and P. californica revealed by fluorescence in situ hybridization. Genome 41(6):848–853
Zhang DM, Sang T (1999) Physical mapping of ribosomal RNA genes in peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: implications for phylogeny and concerted evolution. Am J Bot 86(5):735–740
Acknowledgments
We are grateful to Daming Zhang (Center for Systematic and Evolutionary Botany, Institute of Botany, Beijing, People’s Republic of China) for assistance with finishing the manuscript. This research was supported by National Natural Science Foundation of China (NSFC30860027 and NSFC31260044) and Natural Science Foundation of Jiangxi Province (2009GZN0080).
Author information
Authors and Affiliations
Corresponding author
Additional information
Lingling Xu and Tongjian Li contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, L., Li, T., Liao, L. et al. Reticulate evolution in Ranunculus cantonensis polyploid complex and its allied species. Plant Syst Evol 299, 603–610 (2013). https://doi.org/10.1007/s00606-012-0746-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-012-0746-x