Abstract
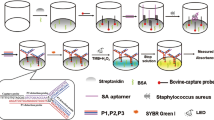
A novel nucleic acid aptamer nanoprobes-mediated hairpin allosteric and aptamer-assisted CRISPR system for detection of Streptococcus pneumoniae and Staphylococcus aureus is presented. In this fluorescence assay system, utilizing the hairpin allosteric effect caused by the aptamer binding to the target bacteria, the detection of S. pneumoniae is first achieved through changes in fluorescence due to FRET. Subsequently, a Cas12a protein mixture is added to detect S. aureus. The amplified output signal is triggered by two methods to ensure the sensitivity of the method: the synergistic FRET effect is achieved by the assembly of multi-aptamer through the conjugation of streptavidin–biotin, and the trans-cleavage function of CRISPR/Cas 12a. Under the optimized conditions, the proposed hairpin allosteric aptasensor could achieve high sensitivity (a detection limit of 135 cfu/mL) and broad-concentration quantification (dynamic range of 103–107 cfu/mL) of S. pneumoniae. The aptamer-assisted CRISPR system for S. aureus detection showed good linearity (R2 = 0.996) in the concentration range 102–108 cfu/mL, with a detection limit of 39 cfu/mL. No cross-reactivity with other foodborne pathogenic bacteria was observed in both systems. Taking only 55 min, this method of multiple pathogen detection proved to be promising.
Graphical Abstract





Similar content being viewed by others
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article [and/or its supplementary materials].
References
Park KS (2018) Nucleic acid aptamer-based methods for diagnosis of infections. Biosens Bioelectron 102:179–188. https://doi.org/10.1016/j.bios.2017.11.028
Hao X, Yeh P, Qin Y, Jiang Y, Qiu Z, Li S et al (2019) Aptamer surface functionalization of microfluidic devices using dendrimers as multi-handled templates and its application in sensitive detections of foodborne pathogenic bacteria. Anal Chim Acta 1056:96–107. https://doi.org/10.1016/j.aca.2019.01.035
Gutiérrez-Santana JC, Toscano-Garibay JD, López-López M, Coria-Jiménez VR (2020) Aptamers coupled to nanoparticles in the diagnosis and treatment of microbial infections. Enfermedades infecciosas y microbiologia clinica (English ed) 38(7):331–337. https://doi.org/10.1016/j.eimc.2019.12.004
Biswas P, Batra S, Gurha N, Maksane N (2022) Emerging antimicrobial resistance and need for antimicrobial stewardship for ocular infections in India: a narrative review. Indian J Ophthalmol 70(5):1513–1521. https://doi.org/10.4103/ijo.IJO_2537_21
Eslami F, Ghasemi Basir HR, Moradi A, Heidari FS (2018) Microbiological study of dacryocystitis in northwest of Iran. Clin Ophthalmol (Auckland, NZ) 12:1859–1864. https://doi.org/10.2147/opth.S175463
Manente R, Santella B, Pagliano P, Santoro E, Casolaro V, Borrelli A et al (2022) Prevalence and antimicrobial resistance of causative agents to ocular infections. Antibiotics (Basel, Switzerland) 11(4). https://doi.org/10.3390/antibiotics11040463
Miller RR, Montoya V, Gardy JL, Patrick DM, Tang P (2013) Metagenomics for pathogen detection in public health. Genome Med 5(9):81. https://doi.org/10.1186/gm485
Zhao X, Li M, Xu Z (2018) Detection of foodborne pathogens by surface enhanced Raman spectroscopy. Front Microbiol 9:1236. https://doi.org/10.3389/fmicb.2018.01236
Mi F, Hu C, Wang Y, Wang L, Peng F, Geng P et al (2022) Recent advancements in microfluidic chip biosensor detection of foodborne pathogenic bacteria: a review. Anal Bioanal Chem 414(9):2883–2902. https://doi.org/10.1007/s00216-021-03872-w
Liu S, Zhao K, Huang M, Zeng M, Deng Y, Li S et al (2022) Research progress on detection techniques for point-of-care testing of foodborne pathogens. Front Bioeng Biotechnol 10:958134. https://doi.org/10.3389/fbioe.2022.958134
Shkembi X, Skouridou V, Svobodova M, Leonardo S, Bashammakh AS, Alyoubi AO et al (2021) Hybrid antibody-aptamer assay for detection of tetrodotoxin in pufferfish. Anal Chem 93(44):14810–14819. https://doi.org/10.1021/acs.analchem.1c03671
Hong L, Pan M, Xie X, Liu K, Yang J, Wang S et al (2021) Aptamer-based fluorescent biosensor for the rapid and sensitive detection of allergens in food matrices. Foods (Basel, Switzerland) 10(11):2598. https://doi.org/10.3390/foods10112598
Buglak AA, Samokhvalov AV, Zherdev AV, Dzantiev BB (2020) Methods and applications of in silico aptamer design and modeling. Int J Mol Sci 21(22):8420. https://doi.org/10.3390/ijms21228420
Stidham S, Villareal V, Chellappa V, Yoder L, Alley O, Shreffler W et al (2022) Aptamer based point of care diagnostic for the detection of food allergens. Sci Rep 12(1):1303. https://doi.org/10.1038/s41598-022-05265-0
Chen W, Lai Q, Zhang Y, Liu Z (2022) Recent advances in aptasensors for rapid and sensitive detection of Staphylococcus aureus. Front Bioeng Biotechnol 10:889431. https://doi.org/10.3389/fbioe.2022.889431
Reich P, Stoltenburg R, Strehlitz B, Frense D, Beckmann D (2017) Development of an impedimetric aptasensor for the detection of Staphylococcus aureus. Int J Mol Sci 18(11). https://doi.org/10.3390/ijms18112484
Shen J, Zhou T, Huang R (2019) Recent advances in electrochemiluminescence sensors for pathogenic bacteria detection. Micromachines 10(8):532. https://doi.org/10.3390/mi10080532
Yu T, Xu H, Zhao Y, Han Y, Zhang Y, Zhang J et al (2020) Aptamer based high throughput colorimetric biosensor for detection of Staphylococcus aureus. Sci Rep 10(1):9190. https://doi.org/10.1038/s41598-020-66105-7
Ma X, Lin X, Xu X, Wang Z (2021) Fabrication of gold/silver nanodimer SERS probes for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Mikrochim Acta 188(6):202. https://doi.org/10.1007/s00604-021-04791-4
Li Z, Zhao W, Ma S, Li Z, Yao Y, Fei T (2021) A chemical-enhanced system for CRISPR-based nucleic acid detection. Biosens Bioelectron 192:113493. https://doi.org/10.1016/j.bios.2021.113493
Zhao G, Wang J, Yao C, Xie P, Li X, Xu Z et al (2022) Alkaline lysis-recombinase polymerase amplification combined with CRISPR/Cas12a assay for the ultrafast visual identification of pork in meat products. Food Chem 383:132318. https://doi.org/10.1016/j.foodchem.2022.132318
Teng J, Yuan F, Ye Y, Zheng L, Yao L, Xue F et al (2016) Aptamer-based technologies in foodborne pathogen detection. Front Microbiol 7:1426. https://doi.org/10.3389/fmicb.2016.01426
Li HY, Jia WN, Li XY, Zhang L, Liu C, Wu J (2020) Advances in detection of infectious agents by aptamer-based technologies. Emerg Microbes Infect 9(1):1671–1681. https://doi.org/10.1080/22221751.2020.1792352
Wang CH, Lee GB (2020) Screening of multiple hemoprotein-specific aptamers and their applications for the binding, quantification, and extraction of hemoproteins in a microfluidic system. Biomicrofluidics 14(2):024110. https://doi.org/10.1063/1.5141871
Yu Y, Han J, Yin J, Huang J, Liu J, Geng L et al (2022) Dual-target electrochemical sensor based on 3D MoS(2)-rGO and aptamer functionalized probes for simultaneous detection of mycotoxins. Front Chem 10:932954. https://doi.org/10.3389/fchem.2022.932954
Yan C, Zhang J, Yao L, Xue F, Lu J, Li B et al (2018) Aptamer-mediated colorimetric method for rapid and sensitive detection of chloramphenicol in food. Food Chem 260:208–212. https://doi.org/10.1016/j.foodchem.2018.04.014
Bilibana MP, Citartan M, Fuku X, Jijana AN, Mathumba P, Iwuoha E (2022) Aptamers functionalized hybrid nanomaterials for algal toxins detection and decontamination in aquatic system: current progress, opportunities, and challenges. Ecotoxicol Environ Saf 232:113249. https://doi.org/10.1016/j.ecoenv.2022.113249
McKeague M (2017) Aptamers for DNA damage and repair. Int J Mol Sci 18(10):2212. https://doi.org/10.3390/ijms18102212
Perez-Gonzalez C, Lafontaine DA, Penedo JC (2016) Fluorescence-based strategies to investigate the structure and dynamics of aptamer-ligand complexes. Front Chem 4:33. https://doi.org/10.3389/fchem.2016.00033
Funding
The authors acknowledge the support from the Key Project of Sichuan Province (No. 22ZDYF1033), the central government guides local science and technology development funds to be directed transfer payment projects (No. 22ZYZF0007), the National Natural Science Foundation of China (No. 2022NSFSC1426), and Wine City Excellence—Science and Technology Innovation Team ([2021] 162).
Author information
Authors and Affiliations
Contributions
Limei Zhang: conceptualization, data curation, writing—original draft, writing—review and editing. Xuejing Xu: writing—review and editing, visualization. Linhong Cao: writing—review and editing. Zixin Zhu: writing—review and editing. Yinhuan Ding: funding support. Hui Jiang: data curation. Baolin Li: writing—review and editing, supervision. Jinbo Liu: funding support, supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Xu, X., Cao, L. et al. Multi-aptamer–mediated hairpin allosteric and aptamer-assisted CRISPR system for detection of S. pneumoniae and S. aureus. Microchim Acta 191, 29 (2024). https://doi.org/10.1007/s00604-023-06094-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-06094-2