Abstract
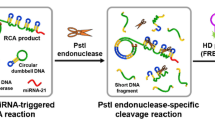
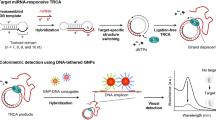
Emerging evidence suggests that exosomal microRNAs are potential biomarkers for the early diagnosis and prognostic assessment of tumor. Here, we design a strand displacement-initiated G-quadruplex/rolling circle amplification (RCA) strategy for highly specific and sensitive electrochemical sensing of exosomal microRNAs. In the presence of exosomal miRNA-21, a locked nucleic acid (LNA)-labeled toehold mediated strand displacement reaction (TMSDR) is initiated, releasing output P2 to trigger the subsequent RCA reaction by hybridizing with the C-rich circular template. Then the obtained G-rich RCA products can bind to the probe anchored on the surface of gold electrode and generate G-quadruplex conformations. Based on the TMSDR-triggered G-quadruplex/RCA strategy, the detection limit of this electrochemical biosensor is down to 2.75 fM. Moreover, our biosensor exhibits excellent repeatability, stability, and high consistency compared to RT-PCR for clinical detection. In conclusion, this assay is expected to provide a hopeful strategy for the early non-invasive diagnosis and prognostic estimation of cancer.

Schematic illustration of electrochemical sensing of exosomal microRNAs based on strand displacement-initiated G-quadruplex/rolling circle amplification (RCA) strategy.







Similar content being viewed by others
References
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A (2013) Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 10:472–484
Sorbara L, Srivastava S (2019) Liquid biopsy: a holy grail for cancer detection. Biomark Med 13:991–994
Hannafon BN, Ding WQ (2013) Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci 14:14240–14269
He X, Ou C (2017) Exosome-derived microRNAs in cancer progression: angel or devil? J Thorac Dis 9:3440–3442
Tsui NB, Ng EK, Lo YM (2002) Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem 48:1647–1653
Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T (2010) Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285:17442–17452
Yoshikawa M, Iinuma H, Umemoto Y, Yanagisawa T, Matsumoto A, Jinno H (2018) Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett 15:9584–9592
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10:42–46
Taylor DD, Gercel-Taylor C (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110:13–21
Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, Gorden P, Kahn CR (2017) Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542:450–455
Ach RA, Wang H, Curry B (2008) Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol 8:69
Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B (2010) A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res 38:e98
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369
Zhang X, Liu C, Pei Y, Song W, Zhang S (2019) Preparation of a novel Raman probe and its application in the detection of circulating tumor cells and Exosomes. ACS Appl Mater Interfaces 11:28671–28680
Wang Y, Li Z, Liu M, Xu J, Hu D, Lin Y, Li J (2017) Multiple-targeted graphene-based nanocarrier for intracellular imaging of mRNAs. Anal Chim Acta 983:1–8
Li XY, Cui YX, Du YC, Tang AN, Kong DM (2019) Label-free telomerase detection in single cell using a Five-Base telomerase product-triggered exponential rolling circle amplification strategy. Acs Sensors 4:1090–1096
Zhou L, Wang Y, Yang C, Xu H, Luo J, Zhang W, Tang X, Yang S, Fu W, Chang K, Chen M (2019) A label-free electrochemical biosensor for microRNAs detection based on DNA nanomaterial by coupling with Y-shaped DNA structure and non-linear hybridization chain reaction. Biosens Bioelectron 126:657–663
Zhao L, Sun R, He P, Zhang X (2019) Ultrasensitive detection of Exosomes by target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical Ratiometric biosensing. Anal Chem 91:14773–14779
Li Y, Teng X, Zhang K, Deng R, Li J (2019) RNA Strand displacement responsive CRISPR/Cas9 system for mRNA sensing. Anal Chem 91:3989–3996
Jiang HX, Liang ZZ, Ma YH, Kong DM, Hong ZY (2016) G-quadruplex fluorescent probe-mediated real-time rolling circle amplification strategy for highly sensitive microRNA detection. Anal Chim Acta 943:114–122
Deng R, Zhang K, Li J (2017) Isothermal amplification for MicroRNA detection: from the test tube to the cell. Acc Chem Res 50:1059–1068
Yi L, Qinli P, Junlong L, Lili Z, Yiyi T, Yuxia L, Wen Y, Guoming X (2017) An "off-on" fluorescent switch assay for microRNA using nonenzymatic ligation-rolling circle amplification. Mikrochim Acta 184:4323
Du YC, Zhu YJ, Li XY, Kong DM (2018) Amplified detection of genome-containing biological targets using terminal deoxynucleotidyl transferase-assisted rolling circle amplification. Chem Commun 54:682–685
Sen D, Gilbert W (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334:364–366
Qian Y, Fan TT, Wang P, Zhang X, Luo JJ, Zhou FY, Yao Y, Liao XJ, Li YH, Gao FL (2017) A novel label-free homogeneous electrochemical immunosensor based on proximity hybridization-triggered isothermal exponential amplification induced G-quadruplex formation. Sensor Actuat B-Chem 248:187–194
Gao FL, Fan TT, Wu J, Liu S, Du Y, Yao Y, Zhou FY, Zhang Y, Liao XJ, Geng DQ (2017) Proximity hybridization triggered hemin/G-quadruplex formation for construction a label-free and signal-on electrochemical DNA sensor. Biosens Bioelectron 96:62–67
Zou R, Zhang F, Chen C, Cai C (2019) An ultrasensitive guanine wire-based resonance light scattering method using G-quadruplex self-assembly for determination of microRNA-122. Mikrochim Acta 186:599
Zhang FT, Nie J, Zhang DW, Chen JT, Zhou YL, Zhang XX (2014) Methylene blue as a G-quadruplex binding probe for label-free homogeneous electrochemical biosensing. Anal Chem 86:9489–9495
Srinivas N, Ouldridge TE, Sulc P, Schaeffer JM, Yurke B, Louis AA, Doye JP, Winfree E (2013) On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res 41:10641–10658
Li S, Liu X, Pang S, Lu R, Liu Y, Fan M, Jia Z, Bai H (2018) Voltammetric determination of DNA based on regulation of DNA strand displacement using an allosteric DNA toehold. Mikrochim Acta 185:433
Chen J, Zhang J, Wang K, Lin X, Huang L, Chen G (2008) Electrochemical biosensor for detection of BCR/ABL fusion gene using locked nucleic acids on 4-aminobenzenesulfonic acid-modified glassy carbon electrode. Anal Chem 80:8028–8034
Wegman DW, Ghasemi F, Stasheuski AS, Khorshidi A, Yang BB, Liu SK, Yousef GM, Krylov SN (2016) Achieving single-nucleotide specificity in direct quantitative analysis of multiple MicroRNAs (DQAMmiR). Anal Chem 88:2472–2477
Wen SW, Sceneay J, Lima LG, Wong CS, Becker M, Krumeich S, Lobb RJ, Castillo V, Wong KN, Ellis S, Parker BS, Moller A (2016) The biodistribution and immune suppressive effects of breast Cancer-derived Exosomes. Cancer Res 76:6816–6827
Yuan YH, Chi BZ, Wen SH, Liang RP, Li ZM, Qiu JD (2018) Ratiometric electrochemical assay for sensitive detecting microRNA based on dual-amplification mechanism of duplex-specific nuclease and hybridization chain reaction. Biosens Bioelectron 102:211–216
Patra S, Roy E, Madhuri R, Sharma PK (2015) Imprinted ZnO nanostructure-based electrochemical sensing of calcitonin: a clinical marker for medullary thyroid carcinoma. Anal Chim Acta 853:271–284
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81430053, 81972027), Chongqing Health Commission (2018QNXM049, 2019ZDXM025) and Medical pre-research project of the Army Medical University (2018XYY04).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1.42 mb)
Rights and permissions
About this article
Cite this article
Tang, X., Wang, Y., Zhou, L. et al. Strand displacement-triggered G-quadruplex/rolling circle amplification strategy for the ultra-sensitive electrochemical sensing of exosomal microRNAs. Microchim Acta 187, 172 (2020). https://doi.org/10.1007/s00604-020-4143-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-4143-9