Abstract
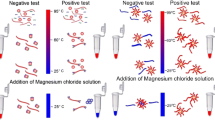
A colorimetric method is presented for the detection of specific nucleotide sequences in plant pathogens. It is based on the use of CRISPR/Cas9-triggered isothermal amplification and gold nanoparticles (AuNPs) as optical probes. The target DNA was recognized and broken up by a given Cas9/sgRNA complex. After isothermal amplification, the product was hybridized with oligonucleotide-functionalized AuNPs. This resulted in the aggregation of AuNPs and a color change from wine red to purple. The visual detection limit is 2 pM of DNA, while a linear relationship exists between the ratio of absorbance at 650 and 525 nm and the DNA concentration in the range from 0.2 pM to 20 nM. In contrast to the previous CRISPR-based amplification platforms, the method has significantly higher specificity with the single-base mismatch and can be visually read out. It was successfully applied to identify the Phytophthora infestans genomic DNA.

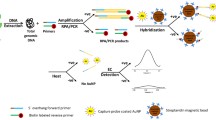
Schematic presentation of a colorimetric method for detection of Phytophthora infestans genomic DNA based on CRISPR/Cas9-triggered isothermal amplification. The Cas9 endonuclease cleaves DNA at the design site and the color changes from red to purple with increasing target DNA concentration.





Similar content being viewed by others
References
Fox A, Mumford RA (2017) Plant viruses and viroids in the United Kingdom: an analysis of first detections and novel discoveries from 1980 to 2014. Virus Res 241:10–18
Khater M, de la Escosura-Muñiz A, Merkoçi A (2017) Biosensors for plant pathogen detection. Biosens Bioelectron 93:72–86
Martinelli F, Scalenghe R, Davino S, Panno S, Scuderi G, Ruisi P, Villa P, Stroppiana D, Boschetti M, Goulart LR, Davis CE, Dandekar AM (2015) Advanced methods of plant disease detection. A review. Agron Sustain Dev 35(1):1–25
Fang Y, Ramasamy RP (2015) Current and prospective methods for plant disease detection. Biosensors 4:537–561
Martín S, Alioto D, Milne RG, Guerri J, Moreno P (2002) Detection of Citrus psorosis virus in field trees by direct tissue blot immunoassay in comparison with ELISA, symptomatology, biological indexing and cross-protection tests. Plant Pathol 51(2):134–141
Shojaei TR, Salleh MAM, Sijam K, Rahim RA, Mohsenifar A, Safarnejad R, Tabatabaei M (2016) Fluorometric immunoassay for detecting the plant virus Citrus tristeza using carbon nanoparticles acting as quenchers and antibodies labeled with CdTe quantum dots. Microchim Acta 183:2277–2287
Toh SY, Citartan M, Gopinath SCB, Tang TH (2015) Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron 64(15):392–403
Babu BK, Sharma R (2015) TaqMan real-time PCR assay for the detection and quantification of Sclerospora graminicola, the causal agent of pearl millet downy mildew. Eur J Plant Pathol 142(1):149–158
De Sousa MV, Machado J d C, Simmons HE, Munkvold GP (2015) Real-time quantitative PCR assays for the rapid detection and quantification of Fusarium oxysporum f. sp. phaseoli in Phaseolus vulgaris (common bean) seeds. Plant Pathol 64(2):478–488
Osman F, Hodzic E, Kwon SJ, Wang J, Vidalakis G (2015) Development and validation of a multiplex reverse transcription quantitative PCR (RT-qPCR) assay for the rapid detection of Citrus tristeza virus, Citrus psorosis virus, and Citrus leaf blotch virus. J Virol Methods 220(3):64–75
Yang L, Tao Y, Yue G, Li R, Qiu B, Guo L, Lin Z, Yang HH (2016) Highly selective and sensitive electrochemiluminescence biosensor for p53 DNA sequence based on nicking endonuclease assisted target recycling and hyperbranched rolling circle amplification. Anal Chem 88(10):5097–5103
Kil EJ, Kim S, Lee YJ, Kang EH, Lee M, Cho SH, Kim MK, Lee KY, Heo NY, Choi HS, Kwon ST, Lee S (2015) Advanced loop-mediated isothermal amplification method for sensitive and specific detection of Tomato chlorosis virus using a uracil DNA glycosylase to control carry-over contamination. J Virol Methods 213:68–74
Kong C, Wang Y, Fodjo EK, Yang G, Han F, Shen X (2018) Loop-mediated isothermal amplification for visual detection of Vibrio parahaemolyticus using gold nanoparticles. Microchim Acta 185(1):35
Bentsink L, Leone G, Van Beckhoven JRCM, Van Schijndel HB, Van Gemen B, Van der Wolf JM (2002) Amplification of RNA by NASBA allows direct detection of viable cells of Ralstonia solanacearum in potato. J Appl Microbiol 93(4):647–655
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6):1262–1278
Zhang F, Wen Y, Guo X (2014) CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23(R1):R40–R46
Liu W, Yu H, Zhou X, Xing D (2016) In vitro evaluation of CRISPR/Cas9 function by an electrochemiluminescent assay. Anal Chem 88(17):8369–8374
Zhang K, Deng R, Li Y, Zhang L, Li J (2016) Cas9 cleavage assay for pre-screening of sgRNAs using nicking triggered isothermal amplification. Chem Sci 7(8):4951–4957
Huang M, Zhou X, Wang H, Xing D (2018) Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal Chem 90(3):2193–2200
Zhao Y, Chen F, Li Q, Wang L, Fan C (2015) Isothermal amplification of nucleic acids. Chem Rev 115(22):12491–12545
Wang J, Zou B, Rui J, Song Q, Kajiyama T, Kambara H, Zhou G (2015) Exponential amplification of DNA with very low background using graphene oxide and single-stranded binding protein to suppress non-specific amplification. Microchim Acta 182:1095–1101
Piao J, Zhou X, Wu X (2018) Colorimetric human papillomavirus DNA assay based on the retardation of avidin-induced aggregation of gold nanoparticles. Microchim Acta 185(12):537
Ma X, Guo Z, Mao Z, Tang Y, Miao P (2018) Colorimetric theophylline aggregation assay using an RNA aptamer and non-crosslinking gold nanoparticles. Microchim Acta 185(1):33
Hu B, Guo J, Xu Y, Wei H, Zhao G, Guan Y (2017) A sensitive colorimetric assay system for nucleic acid detection based on isothermal signal amplification technology. Anal Bioanal Chem 409(20):4819–4825
Liu J (2012) Adsorption of DNA onto gold nanoparticles and graphene oxide: surface science and applications. Phys Chem Chem Phys 14(30):10485–10496
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529(7587):490–495
Shi C, Liu Q, Zhou M, Zhao H, Yang T, Ma C (2016) Nicking endonuclease-mediated isothermal exponential amplification for double-stranded DNA detection. Sensors Actuators B Chem 222:221–225
Xu SY, Zhu Z, Zhang P, Chan SY, Samuelson JC, Xiao J, Ingalls D, Wilson GG (2007) Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI. Nucleic Acids Res 35(14):4608–4618
Dharanivasan G, Riyaz SUM, Jesse DMI, Muthuramalingam TR, Rajendran G, Kathiravan K (2016) DNA templated self-assembly of gold nanoparticle clusters in the colorimetric detection of plant viral DNA using a gold nanoparticle conjugated bifunctional oligonucleotide probe. RSC Adv 6(14):11773–11785
Deng H, Xu Y, Liu Y, Che Z, Guo H, Shan S, Sun Y, Liu X, Huang K, Ma X, Wu Y, Liang XJ (2012) Gold nanoparticles with asymmetric polymerase chain reaction for colorimetric detection of DNA sequence. Anal Chem 84(3):1253–1258
Sperling RA, Parak WJ (2010) Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Phil Trans R Soc A 368(1915):1333–1383
Yoshitake k WS, Ueda H (2008) Dimerization-based homogeneous fluorosensor proteins for the detection of specific dsDNA. Biosens Bioelectron 23:1266–1271
Qiu L, Shen Z, Wu ZS, Shen GL, Yu R (2015) Discovery of the unique self-assembly behavior of terminal suckerscontained dsDNA onto GNP and novel “light-up” colorimetric assay of nucleic acids. Biosens Bioelectron 64:292–299
Ermini ML, Mariani S, Scarano S, Minunni M (2014) Bioanalytical approaches for the detection of single nucleotide polymorphisms by surface plasmon resonance biosensors. Biosens Bioelectron 61:28–37
Li S, Liu H, Jia Y, Deng Y, Zhang L, Lu Z, He N (2012) A novel SNPs detection method based on gold magnetic nanoparticles array and single base extension. Theranostics 2(10):967–975
Acknowledgments
This work was supported by the National Scientific Foundation of China (21705051, 21874048), the Scientific Foundation of Guangdong Province (2017A030313077), the Science and Technology Planning Project of Guangdong Province (2016B030303010), National Key Research and Development Program of China (SQ2017YFC160089), the Program for the Top Young Innovative Talents of Guangdong Province (2016TQ03N305), and the Foundation for High-level Talents in South China Agricultural University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 986 kb)
Rights and permissions
About this article
Cite this article
Chang, W., Liu, W., Liu, Y. et al. Colorimetric detection of nucleic acid sequences in plant pathogens based on CRISPR/Cas9 triggered signal amplification. Microchim Acta 186, 243 (2019). https://doi.org/10.1007/s00604-019-3348-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3348-2