Abstract
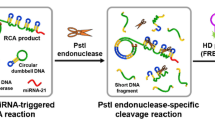
The authors present a fluorometric method for ultrasensitive determination of the activity of uracil-DNA glycosylase (UDG). It is based on the use of two-tailed reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and an entropy-driven reaction. The assay involves the following steps: (1) UDG-driven uracil excision repair, (2) two-tailed RT-qPCR-mediated amplification, (3) RNA polymerase-aided amplification, and (4) DNA-modified silver nanoclusters (AgNCs) as a transducer to produce a fluorescent signal. UDG enables uracil to be removed from U·A pairs in DNA1 and produces a depurinated/depyrimidinated site that is readily cleaved by endonuclease IV (Endo IV). The cleaved DNA contains the T7 RNA polymerase primer for the T7 RNA polymerase amplification which produces a large number of microRNA sequences. Subsequent two-tailed RT-qPCR leads to the formation of a prolonged DNA termed DNA3. The prolonged part of DNA3 is then hybridized with an added DNA4/DNA5 duplex, where DNA5 is labeled with gold nanoparticles (AuNPs), and DNA 4 is labeled with AgNCs. The AuNPs quench the fluorescence of the AgNCs. The duplex has a toehold to hybridize the prolong part of DNA3. This results in the formation of a DNA5/DNA3 duplex due to strand displacement (by replacing the DNA4 in the DNA4/DNA5 duplex). DNA4 is released and moves away from the AuNPs. This results in restored AgNC fluorescence, best measured at excitation/emission wavelengths of 575/635 nm. The method has a detection limit as low as 0.1 mU mL−1 of UDG activity (3σ criterion) with a range of 0.001–0.01 U mL−1. It was used to measure UDG activity in cell lysates. Conceivably, it may be used to screen for UDG inhibitors such as Ugi.

Schematic presentation of the two-tailed RT-qPCR assay platform for ultrasensitive detection of uracil-DNA glycosylase (UDG). Two-tailed RT-qPCR-mediated amplification and RNA polymerase-aided amplification are utilized for signal amplification. DNA-modified silver nanoclusters (AgNCs) are used as a transducer to produce a fluorescent signal.






Similar content being viewed by others
References
Marnett LJ, Plastaras JP (2001) Endogenous DNA damage and mutation. Trends Genet 17(4):214–221
Tubbs A, Nussenzweig A (2017) Endogenous DNA damage as a source of genomic instability in Cancer. Cell 168(4):644–656
Wang Y, Wang Y, Li D, Xu J, Ye C (2018) Detection of nucleic acids and elimination of carryover contamination by using loop-mediated isothermal amplification and antarctic thermal sensitive uracil-DNA-glycosylase in a lateral flow biosensor: application to the detection of Streptococcus pneumoni. Microchim Acta 185(4):212
Prorok P, Alili D, Saint-Pierre C, Gasparutto D, Zharkov DO, Ishchenko AA, Tudek B, Saparbaev MK (2013) Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Pnas 110(39):E3695–E3703
Sousa MM, Krokan HE, Slupphaug G (2007) DNA-uracil and human pathology. Mol Asp Med 28(3):276–306
Du Y-C, Cui Y-X, Li X-Y, Sun G-Y, Zhang Y-P, Tang A-N, Kim K, Kong D-M (2018) Terminal Deoxynucleotidyl transferase and T7 exonuclease-aided amplification strategy for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem 90(14):8629–8634
Mcwilliams MA, Anka FH, Balkus KJ, Slinker JD (2014) Sensitive and selective real-time electrochemical monitoring of DNA repair. Biosens Bioelectron 54(54C):541–546
Liu B, Yang X, Wang K, Tan W, Li H, Tang H (2007) Real-time monitoring of uracil removal by uracil-DNA glycosylase using fluorescent resonance energy transfer probes. Anal Biochem 366(2):237–243
Wu Y, Wang L, Zhu J, Jiang W (2015) A DNA machine-based fluorescence amplification strategy for sensitive detection of uracil-DNA glycosylase activity. Biosens Bioelectron 68:654–659
Dong J, Lian J, Jin Y, Li B (2017) Guanine-based chemiluminescence resonance energy transfer biosensing platform for the specific assay of uracil-DNA glycosylase activity. Anal Methods 9(2):276–281
Tao Y, Wei Q (2018) Plasmonic molecular assays: recent advances and applications for mobile health. Nano Res:1–35
Viturro E, Altenhofer C, Zölch B, Burgmaier A, Riedmaier I, Pfaffl MW (2014) Microfluidic high-throughput reverse-transcription quantitative PCR analysis of liver gene expression in lactating animals. Microchim Acta 181(13–14):1725–1732
Pinho R, Guedes LC, Soreq L, Lobo PP, Mestre T, Coelho M, Rosa MM, Gonçalves N, Wales P, Mendes T (2016) Correction: Gene Expression Differences in Peripheral Blood of Parkinson's Disease Patients with Distinct Progression Profiles. Plos One 11(6):e0157852
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39(4):519–525
Androvic P, Valihrach L, Elling J, Sjoback R, Kubista M (2017) Two-tailed RT-qPCR: a novel method for highly accurate miRNA quantification. Nucleic Acids Res 45(15):e144
Zhang DY, Turberfield AJ, Yurke B, Winfree E (2007) Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318(5853):1121–1125
Lv Y, Cui L, Peng R, Zhao Z, Qiu L, Chen H, Jin C, Zhang XB, Tan W (2015) Entropy Beacon: a hairpin-free DNA amplification strategy for efficient detection of nucleic acids. Anal Chem 87(23):11714–11720
He X, Zeng T, Li Z, Wang G, Ma N (2016) Catalytic molecular imaging of MicroRNA in living cells by DNA-programmed nanoparticle disassembly. Angew Chem Int Ed 55(9):3073–3076
Zhou H, Liu J, Xu J-J, Zhang S-S, Chen H-Y (2018) Optical nano-biosensing interface nucleic acid amplification strategy: construction and application. Chem Soc Rev 47(6):1996–2019
Zhang K, Yang X-J, Zhao W, Xu M-C, Xu J-J, Chen H-Y (2017) Regulation and imaging of gene expression via an RNA interference antagonistic biomimetic probe. Chem Sci 8(7):4973–4977
Liu J, Cui M, Niu L, Zhou H, Zhang S (2016) Enhanced peroxidase-like properties of graphene-hemin-composite decorated with Au nanoflowers as electrochemical aptamer biosensor for the detection of K562 leukemia cancer cells. Chem Eur J 22(50):18001–18008
Zhang K, Wang K, Zhu X, Gao Y, Xie M (2014) Rational design of signal-on biosensors by using photoinduced electron transfer between ag nanoclusters and split G-quadruplex halves-hemin complexes. Chem Commun 50(91):14221–14224
Zhang K, Wang K, Xie M, Zhu X, Xu L, Yang R, Huang B, Zhu X (2014) DNA-templated silver nanoclusters based label-free fluorescent molecular beacon for the detection of adenosine deaminase. Biosens Bioelectron 52(0):124–128
Zhang K, Wang K, Zhu X, Xie M (2017) Ultrasensitive fluorescence detection of transcription factors based on kisscomplex formation and the T7 RNA polymerase amplification method. Chem Commun 53(43):5846–5849
Wang LJ, Ren M, Zhang Q, Tang B, Zhang CY (2017) Excision repair-initiated enzyme-assisted bicyclic Cascade signal amplification for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem 89(8):4488–4494
Zhu J, Hao Q, Liu Y, Guo Z, Rustam B, Jiang W (2018) Integrating DNA structure switch with branched hairpins for the detection of uracil-DNA glycosylase activity and inhibitor screening. Talanta 179:51–56
Zhang Y, Li QN, Li CC, Zhang CY (2018) Label-free and high-throughput bioluminescence detection of uracil-DNA glycosylase in cancer cells through tricyclic cascade signal amplification. Chem Commun (Camb) 54(51):6991–6994
Zhang XF, Li N, Ling Y, Li NB, Luo HQ (2018) Sensitive detection of active uracil-DNA glycosylase via an exonuclease III-assisted cascade multi-amplification fluorescence DNA machine. Sensors Actuators B Chem 271:9–14
Zhang X, Wu T, Wang H, Zou Y, Chen W, Zhao M, Wang S, Xiao X (2018) A time-dependent fluorescent biosensor for uracil-DNA glycosylase detection based on the uracil inhibition effect towards archaebacterial DNA polymerases. Sensors Actuators B Chem 270:277–282
Ren R, Shi K, Yang J, Yuan R, Xiang Y (2018) DNA three way junction-mediated recycling amplification for sensitive electrochemical monitoring of uracil-DNA glycosylase activity and inhibition. Sensors Actuators B Chem 258:783–788
Xu X, Wang L, Wu Y, Jiang W (2017) Uracil removal-inhibited ligase reaction in combination with catalytic hairpin assembly for the sensitive and specific detection of uracil-DNA glycosylase activity. Analyst 142(24):4655–4660
Du W, Li J, Xiao F, Yu R, Jiang J (2017) A label-free and highly sensitive strategy for uracil-DNA glycosylase activity detection based on stem-loop primer-mediated exponential amplification (SPEA). Anal Chim Acta 991:127–132
Jiao F, Qian P, Qin Y, Xia Y, Deng C, Nie Z (2016) A novel and label-free biosensors for uracil-DNA glycosylase activity based on the electrochemical oxidation of guanine bases at the graphene modified electrode. Talanta 147:98–102
Nie H, Wang W, Li W, Nie Z, Yao S (2015) A colorimetric and smartphone readable method for uracil-DNA glycosylase detection based on the target-triggered formation of G-quadruplex. Analyst 140(8):2771–2777
Lu YJ, Hu DP, Deng Q, Wang ZY, Huang BH, Fang YX, Zhang K, Wong WL (2015) Sensitive and selective detection of uracil-DNA glycosylase activity with a new pyridinium luminescent switch-on molecular probe. Analyst 140(17):5998–6004
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21705061), the Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017) and the Innovation Capacity Development Plan of Jiangsu Province (BM2018023).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, K., Huang, W., Huang, Y. et al. Determination of the activity of uracil-DNA glycosylase by using two-tailed reverse transcription PCR and gold nanoparticle-mediated silver nanocluster fluorescence: a new method for gene therapy-related enzyme detection. Microchim Acta 186, 181 (2019). https://doi.org/10.1007/s00604-019-3307-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3307-y