Abstract
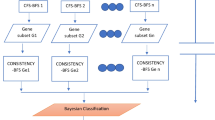
In this study, we aimed to classify bladder cancer patients using tumoral and non-tumoral gene expression data. In this way, we aimed to determine which genes are effective on tumoral and normal tissues. In addition, for this purpose, we planned to perform this classification using interpretable methods (The aim of this study was to classify bladder cancer patients using gene expression data from tumoral and non-tumoral tissues. By doing so, we wanted to determine which genes were effective on both tumoral and normal tissues. Moreover, for this purpose, we planned to use interpretable methods for this classification.). Analyses using permutation feature importance (PFI), SHapley Additive exPlanations (SHAP), local interpretable model-agnostic explanations (LIME), and Anchor methods on data from Gene Expression Omnibus (GEO) and Curated Microarray Database we did (We performed analyses using permutation feature importance (PFI), SHapley Additive exPlanations (SHAP), local interpretable model-agnostic explanations (LIME), and Anchor methods on data from Gene Expression Omnibus (GEO) and Curated Microarray Database.). These are eXplainable methods used to determine the importance of genes in classification. According to the results of our study, the most important genes were determined as LINC00161, ACACB, and CBARP according to the PFI method, HSPA6, STON2, and RFC2 according to the SHAP method, PRUNE2 and ABCC13 according to the LIME method, and TMEM74, KLHL10, and GAMT according to the Anchor method. This study shows that genes involved in other cancer types are also effective in bladder cancer. In addition, it has been observed that using explainable methods in cancer data can support prognosis and treatment in the clinic.






Similar content being viewed by others
Data availability
Data openly available in a public repository (https://www.ncbi.nlm.nih.gov/geo/), (https://sbcb.inf.ufrgs.br/cumida).
References
WHO. Bladder cancer. https://www.iarc.who.int/cancer-type/bladder-cancer/ (accessed 2023).
Segundo-Val IS, Sanz-Lozano CS (2016) Introduction to the gene expression analysis. Methods Mol Biol 1434:29–43. https://doi.org/10.1007/978-1-4939-3652-6_3
Vadapalli S, Abdelhalim H, Zeeshan S, Ahmed Z (2022) Artificial intelligence and machine learning approaches using gene expression and variant data for personalized medicine. Brief Bioinf 23(5):191. https://doi.org/10.1093/bib/bbac191
Abbas M, El-Manzalawy Y (2020) Machine learning based refined differential gene expression analysis of pediatric sepsis. BMC Med ical Genom 13(1):122. https://doi.org/10.1186/s12920-020-00771-4
Guneri-Sozeri PY, Erkek-Ozhan S (2022) Identification of the gene expression changes and gene regulatory aspects in ELF3 mutant bladder cancer. Mol Biol Rep 49(4):3135–3147. https://doi.org/10.1007/s11033-022-07145-2
Zaravinos A, Lambrou GI, Volanis D, Delakas D, Spandidos DA (2011) Spotlight on differentially expressed genes in urinary bladder cancer. PLoS ONE 6(4):e18255. https://doi.org/10.1371/journal.pone.0018255
Khalsan M et al (2022) A survey of machine learning approaches applied to gene expression analysis for cancer prediction. IEEE Access 10:27522–27534. https://doi.org/10.1109/ACCESS.2022.3146312
Rukhsar L, Bangyal WH, Ali Khan MS, Ag Ibrahim AA, Nisar K, Rawat DB (2022) Analyzing RNA-Seq gene expression data using deep learning approaches for cancer classification. Appl Sci 12(4):1850. https://doi.org/10.3390/app12041850
Almarzouki HZ (2022) Deep-learning-based cancer profiles classification using gene expression data profile. J Healthcare Eng 2022:4715998. https://doi.org/10.1155/2022/4715998
Chen K et al (2021) Identification and validation of hub genes associated with bladder cancer by integrated bioinformatics and experimental assays. Front Oncol Original Res 11:782981. https://doi.org/10.3389/fonc.2021.782981
Wagner A (2022) AI predicts the effectiveness and evolution of gene promoter sequences. Nature 603:384. https://doi.org/10.1038/d41586-022-00384-0
Abbod MFMF et al. (2006) Artificial intelligence technique for gene expression profiling of urinary bladder cancer. In: 2006 3rd International IEEE conference intelligent systems, 4–6 Sept 2006, pp 646–651. https://doi.org/10.1109/IS.2006.348495
Altmann A, Toloşi L, Sander O, Lengauer T (2010) Permutation importance: a corrected feature importance measure. Bioinformatics 26(10):1340–1347
Li J et al (2023) Identification of genes related to immune enhancement caused by heterologous ChAdOx1-BNT162b2 vaccines in lymphocytes at single-cell resolution with machine learning methods. Front Immunol 14:1131051. https://doi.org/10.3389/fimmu.2023.1131051
Shew M et al (2021) MicroRNA profiling as a methodology to diagnose Ménière’s disease: potential application of machine learning. Otolaryngol Head Neck Surg 164(2):399–406. https://doi.org/10.1177/0194599820940649
Bazaga A, Leggate D, Weisser H (2020) Genome-wide investigation of gene-cancer associations for the prediction of novel therapeutic targets in oncology. Sci Rep 10(1):10787. https://doi.org/10.1038/s41598-020-67846-1
Shapley L (1953) A value for n-person games. Princeton University Press, Princeton, pp 307–317. https://doi.org/10.1515/9781400881970-018
Derks J, Peters H (1993) A shapley value for games with restricted coalitions. Int J Game Theory 21(4):351–60. Available: https://EconPapers.repec.org/RePEc:spr:jogath:v:21:y:1993:i:4:p:351-60.
Sanchez K, Kamal K, Manjaly P, Ly S, Mostaghimi A (2023) Clinical application of artificial intelligence for non-melanoma skin cancer. Current Treatment Options Oncol 24(4):373–379. https://doi.org/10.1007/s11864-023-01065-4
Kumar S, Das A (2023) Peripheral blood mononuclear cell derived biomarker detection using eXplainable Artificial Intelligence (XAI) provides better diagnosis of breast cancer. Comput Biol Chem 104:107867. https://doi.org/10.1016/j.compbiolchem.2023.107867
Zhu K et al (2022) A novel 10-gene ferroptosis-related prognostic signature in acute myeloid leukemia. Front Oncol 12:1023040. https://doi.org/10.3389/fonc.2022.1023040
Palatnik de Sousa I, Maria Bernardes Rebuzzi Vellasco M, Costa da Silva E (2019) Local interpretable model-agnostic explanations for classification of lymph node metastases. Sensors 19(13), 2969. Available: https://www.mdpi.com/1424-8220/19/13/2969
Lai Y et al (2022) Identification of immune microenvironment subtypes and signature genes for Alzheimer’s disease diagnosis and risk prediction based on explainable machine learning. Front Immunol 13:1046410. https://doi.org/10.3389/fimmu.2022.1046410
Oni O, Qiao S (2019) Model-agnostic interpretation of cancer classification with multi-platform genomic data, pp 34–41
Modhukur V et al. (2021) Machine learning approaches to classify primary and metastatic cancers using tissue of origin-based DNA methylation profiles. Cancers (Basel) 13(15):3768. Available: https://www.mdpi.com/2072-6694/13/15/3768
Marco Tulio Ribeiro SS, Guestrin C (2018) Anchors: high-precision model-agnostic explanations. . Available: https://homes.cs.washington.edu/~marcotcr/aaai18.pdf
Edgar R, Domrachev M, Lash AE (2002) "Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210. https://doi.org/10.1093/nar/30.1.207
Feltes BC, Chandelier E, Grisci B, Dorn M (2019) CuMiDa: an extensively curated microarray database for benchmarking and testing of machine learning approaches in cancer research. J Comput Biol 26. https://doi.org/10.1089/cmb.2018.0238
Sherman BT et al (2022) DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50(W1):W216-w221. https://doi.org/10.1093/nar/gkac194
Botchkarev A (2018) Performance metrics (error measures) in machine learning regression. Forecast Prognost Prop Typol
Vujovic ZD (2021) Classification model evaluation metrics. Int J Adv Comput Sci Appl 12(6):599–606
De Diego IM, Redondo AR, Fernandez RR, Navarro J, Moguerza JM (2022) General performance score for classification problems. Appl Intell 52(10):12049–12063. https://doi.org/10.1007/s10489-021-03041-7
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/a:1010933404324
Octaviani TL, Rustam Z (2019) Random forest for breast cancer prediction. In: 4th International symposium on current progress in mathematics and sciences (ISCPMS). Univ Indonesia, Fac Math and Nat Sci, Depok, INDONESIA, vol 2168. In: AIP Conference Proceedings,30–31 Oct 2018. https://doi.org/10.1063/1.5132477. Available: <Go to ISI>://WOS:000519032600050
Huljanah M, Rustam Z, Utama S, Siswantining T, Iop (2019) Feature selection using random forest classifier for predicting prostate cancer. In: presented at the 9TH annual basic science international conference 2019 (BASIC 2019)
Huang M et al (2017) Head and neck cancer survival outcome prediction based on NRG oncology RTOG 0522 with random forests and random survival forests. Med Phys 44(6)
Liu DF et al (2021) Optimisation and evaluation of the random forest model in the efficacy prediction of chemoradiotherapy for advanced cervical cancer based on radiomics signature from high-resolution T2 weighted images. Arch Gynecol Obstetrics 303(3):811–820. https://doi.org/10.1007/s00404-020-05908-5.
Santhanam R, Uzir N, Raman S, Banerjee S (2017) Experimenting XGBoost algorithm for prediction and classification of different datasets
Deng XS, Li M, Deng SB, Wang L (2022) Hybrid gene selection approach using XGBoost and multi-objective genetic algorithm for cancer classification. Med Biol Eng Comput 60(3):663–681. https://doi.org/10.1007/s11517-021-02476-x
Ma BS et al (2022) Diagnostic classification of cancers using DNA methylation of paracancerous tissues. Sci Rep 12(1):10646. https://doi.org/10.1038/s41598-022-14786-7
Song YY, Lu Y (2015) Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry 27(2):130–135. https://doi.org/10.11919/j.issn.1002-0829.215044
Zhang Z (2016) Introduction to machine learning: k-nearest neighbors. Ann Transl Med 4(11):218. https://doi.org/10.21037/atm.2016.03.37
Lee WM (2019) Supervised learning—classification using K‐nearest neighbors (KNN), pp 205–220
Momodu A (2017) K-nearest neighbor implementation in python 3.6.1 from scratch
Gao S, Li HM (2012) IEEE breast cancer diagnosis based on support vector machine. In: Presented at the 2012 2nd international conference on uncertainty reasoning and knowledge engineering (URKE)
Chen LY, Li JT, Chang MM (2020) Cancer diagnosis and disease gene identification via statistical machine learning. Curr Bioinform 15(9):956–962. https://doi.org/10.2174/1574893615666200207094947
Teeyapan K, Theera-Umpon N, Auephanwiriyakul S, IEEE (2015) Application of support vector based methods for cervical cancer cell classification. In: Presented at the proceedings 5th IEEE international conference on control system, computing and engineering (ICCSCE 2015)
Liu TB, Zhang XM, Chen R, Deng XX, Fu B (2023) Development, comparison, and validation of four intelligent, practical machine learning models for patients with prostate-specific antigen in the gray zone. Front Oncol 13. Art no. 1157384. https://doi.org/10.3389/fonc.2023.1157384
Akcay M, Etiz D, Celik O, Ozen A (2022) Evaluation of acute hematological toxicity by machine learning in gynecologic cancers using postoperative radiotherapy. Indian J Cancer 59(2):178–186. https://doi.org/10.4103/ijc.IJC_666_19
Lei L, IEEE (2018) Research on logistic regression algorithm of breast cancer diagnose data by machine learning. In: presented at the 2018 international conference on robots and intelligent system (ICRIS 2018)
Ramirez SG, Hales RC, Williams GP, Jones NL (2022) Extending SC-PDSI-PM with neural network regression using GLDAS data and Permutation Feature Importance. Environ Model Softw 157:105475
Gramegna A, Giudici P (2021) SHAP and LIME: an evaluation of discriminative power in credit risk. Front Artif Intell 4. Art no. 752558. https://doi.org/10.3389/frai.2021.752558
Holzinger A, Saranti A, Molnar C, Biecek P, Samek W (2022) Explainable AI methods—a brief overview. Springer International Publishing, pp 13–38
Hagras H (2018) Toward human-understandable, explainable AI. Computer 51(9):28–36
Shi Y, Zhou Y (2010) The role of surgery in the treatment of gastric cancer. J Surg Oncol 101(8):687–692. https://doi.org/10.1002/jso.21455
Wilusz JE, Sunwoo H, Spector DL (2009) Long non-coding RNAs: functional surprises from the RNA world. Genes Dev 23(13):1494–1504. https://doi.org/10.1101/gad.1800909
Shen Y et al (2015) Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget 6(11):8579–8592. https://doi.org/10.18632/oncotarget.3287
Sun J et al (2015) A potential prognostic long non-coding RNA signature to predict metastasis-free survival of breast cancer patients. Sci Rep 5(1):16553. https://doi.org/10.1038/srep16553
Li J et al (2014) LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 63(11):1700–1710. https://doi.org/10.1136/gutjnl-2013-305806
Hu Y et al (2014) A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 5(8):2230–2242. https://doi.org/10.18632/oncotarget.1895
Zhou M et al (2015) A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Trans Med 13(1):231. https://doi.org/10.1186/s12967-015-0556-3
Zhou M et al (2016) Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget 7(22):32433–32448. https://doi.org/10.18632/oncotarget.8653
Xu LC et al (2017) Up-regulation of LINC00161 correlates with tumor migration and invasion and poor prognosis of patients with hepatocellular carcinoma. Oncotarget 8(34):56168–56173. https://doi.org/10.18632/oncotarget.17040
Li Z, Dou P, Liu T, He S (2017) Application of long non-coding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem 42(4):1407–1419. https://doi.org/10.1159/000479205
Wang Y et al (2016) Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett 382(2):137–146. https://doi.org/10.1016/j.canlet.2016.08.024
Shin SS et al (2017) HSPA6 augments garlic extract-induced inhibition of proliferation, migration, and invasion of bladder cancer EJ cells; Implication for cell cycle dysregulation, signaling pathway alteration, and transcription factor-associated MMP-9 regulation. PLoS ONE 12(2):e0171860. https://doi.org/10.1371/journal.pone.0171860
Salameh A et al (2015) PRUNE2 is a human prostate cancer suppressor regulated by the intronic long non-coding RNA PCA3. Proc Natl Acad Sci USA 112(27):8403–8408. https://doi.org/10.1073/pnas.1507882112
Zhou C, Li AH, Liu S, Sun H () Identification of an 11-autophagy-related-gene signature as promising prognostic biomarker for bladder cancer patients. Biology (Basel) 10(5). https://doi.org/10.3390/biology10050375
Sun Y et al (2017) TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis 8(8):e3031. https://doi.org/10.1038/cddis.2017.370
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kırboğa, K.K. Bladder cancer gene expression prediction with explainable algorithms. Neural Comput & Applic 36, 1585–1597 (2024). https://doi.org/10.1007/s00521-023-09142-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-09142-3